|
Home |
Download |
Statistics |
Help |
Contact |
Statistics
 Overview of PCTfuncDB.
Overview of PCTfuncDB.We collected 62 single-cell RNA-sequencing (scRNA-seq) and 12 whole exome sequencing (WES) datasets across 6 different platforms from 7 public databases. For scRNA-seq data, after removing low-quality cells and samples, a total of 4,972,145 cells and 1,352 patient samples from 13 tissues with 46 different disease states were retained in PCTanno. To identify malignant transformation related genes, we first catalog epithelial, immune and stromal cell types for each scRNA-seq dataset. Then, we integrate corresponding single-cell transcriptomes for three major cell types above within each tissue, respectively and run batch correction. We also carried out functional impact prediction of mutations in our identified malignant transformation related genes based on bulk DNA and scRNA-seq samples.
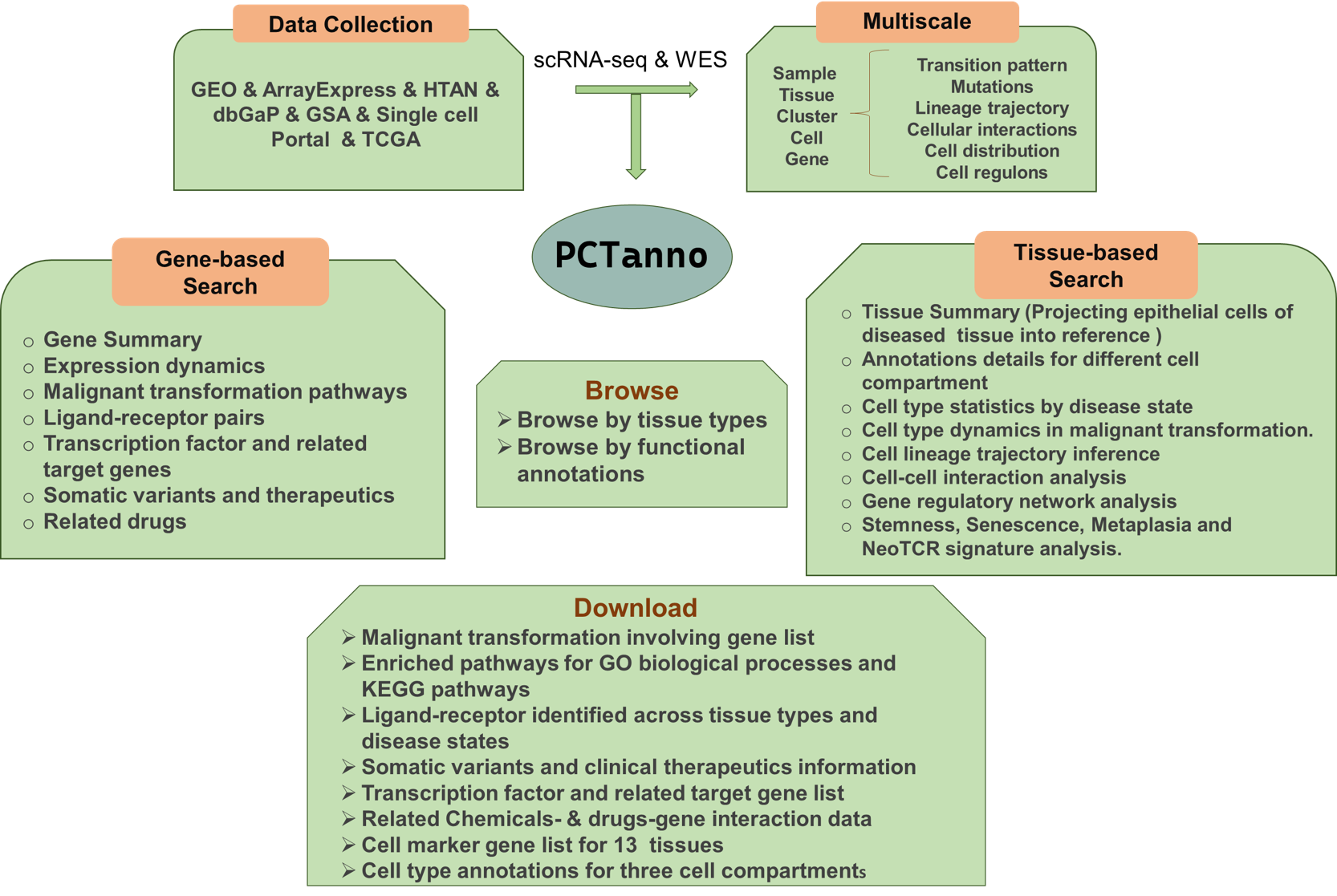
Figure 1. Overview of PCTanno functional annotation.
 Function of PCTanno.
Function of PCTanno.By looking at malignancies through the lens of their originating lesions provide, PCTanno can provide comprehensive characterizations for genes involved in malignant transformation across diverse tissue types in human, and chart cell composition and cell state changes that occur during the transformation of healthy tissues to precancer to cancer.
 Summary of data design and collection for malignant transformation analysis.
Summary of data design and collection for malignant transformation analysis.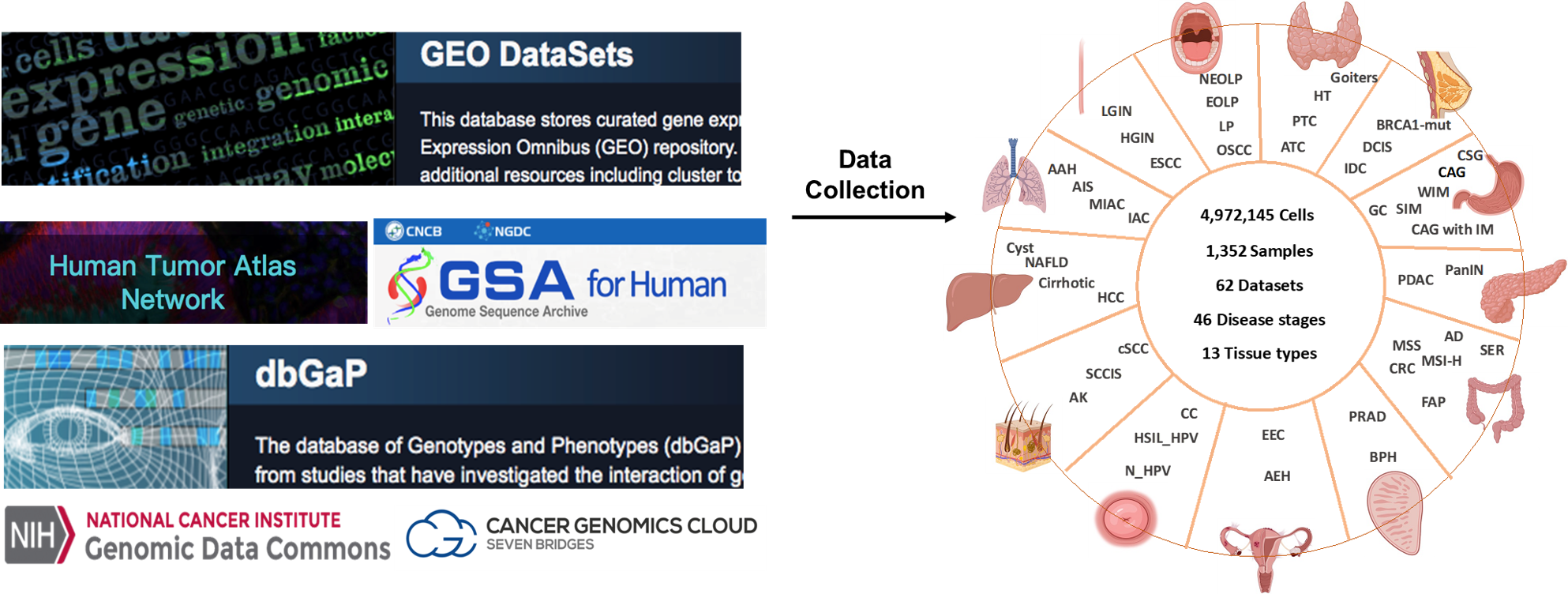
 Summary of the number of cells, samples collected by tissue (left) and their tissue compositions (right).
Summary of the number of cells, samples collected by tissue (left) and their tissue compositions (right).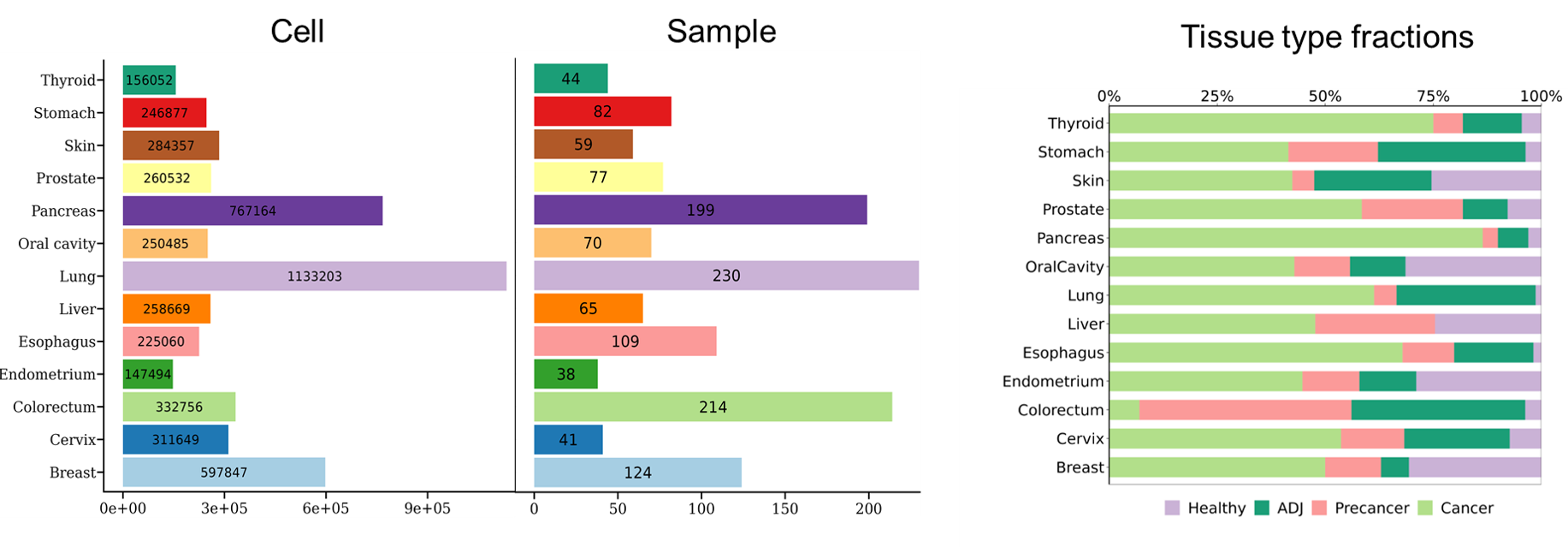
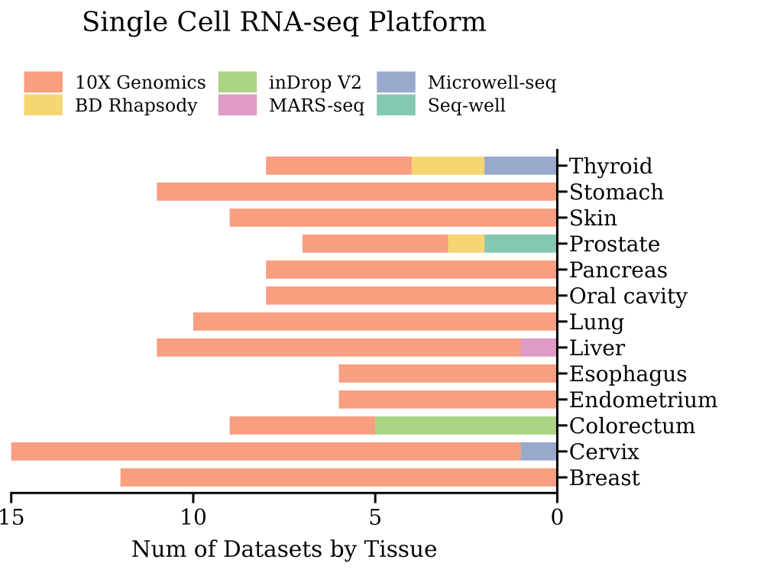
 Summary of platforms of the included scRNA-seq datasets.
Summary of platforms of the included scRNA-seq datasets.