|
Home |
Download |
Statistics |
Help |
Contact |
Instructions for Navigating the Site
1. PCTanno information2. Understanding gene annotation of PCTanno
a. Search by Gene symbol or Entrez gene ID, example: Gene symbol: HNF4A, Entrez gene ID: 3172
b. Gene search result page
c. Gene annotation result page
i. Gene summary
ii. Malignant transformation analysis
iii. Malignant transformation related pathway analysis
iv. Cell-cell communication analysis
v. Single-cell gene regulatory network inference analysis
vi. Somatic mutation of malignant transformation related genes
vii. Related drugs of malignant transformation related genes
3. Understanding tissue annotation of PCTanno
a. Search by tissue, example: Liver
b. Tissue annotation result page
i. Tissue samples summary
ii. Annotation details for different cell compartments
iii. Cell type statistics by disease state
iv. Cell type dynamics in malignant transformation
v. Cell lineage trajectory inference
vi. TF regulatory network analysis
vii. Cell-cell interaction analysis
viii. Stemness, Senescence, Metaplasia and NeoTCR signature analysis
4. Download data and contact us
We analyzed 2 datasets from 52 single cell RNA sequencing (scRNA-seq) studies and 12 whole exome sequencing (WES) datasets that across 12 tissues from healthy to premalignant lesion to cancer.
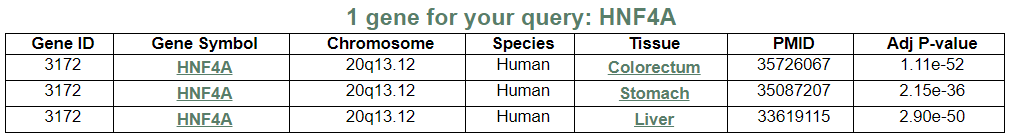
- Input query: Gene symbol or Entrez gene ID.

This image shows the gene list according to your search.
This image shows PCTanno's annotation categories for your query with links to their corresponding annotation parts.

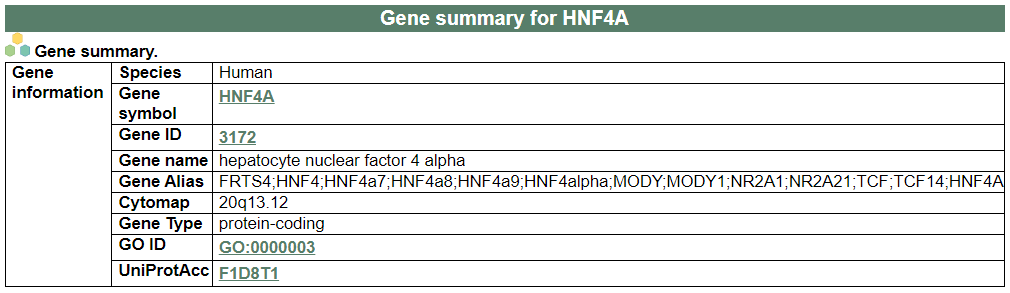
 i. 'Gene summary' category
i. 'Gene summary' categoryThis image category shows the information of the PCTanno.
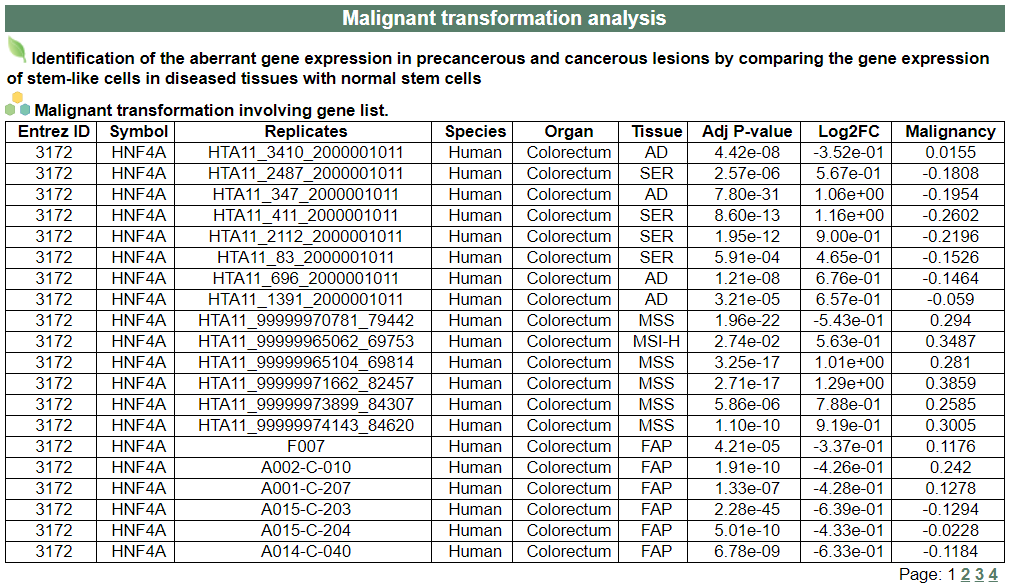
 ii. 'Malignant transformation analysis' category
ii. 'Malignant transformation analysis' categoryThis image category provides the result of Malignant transformation analysis.
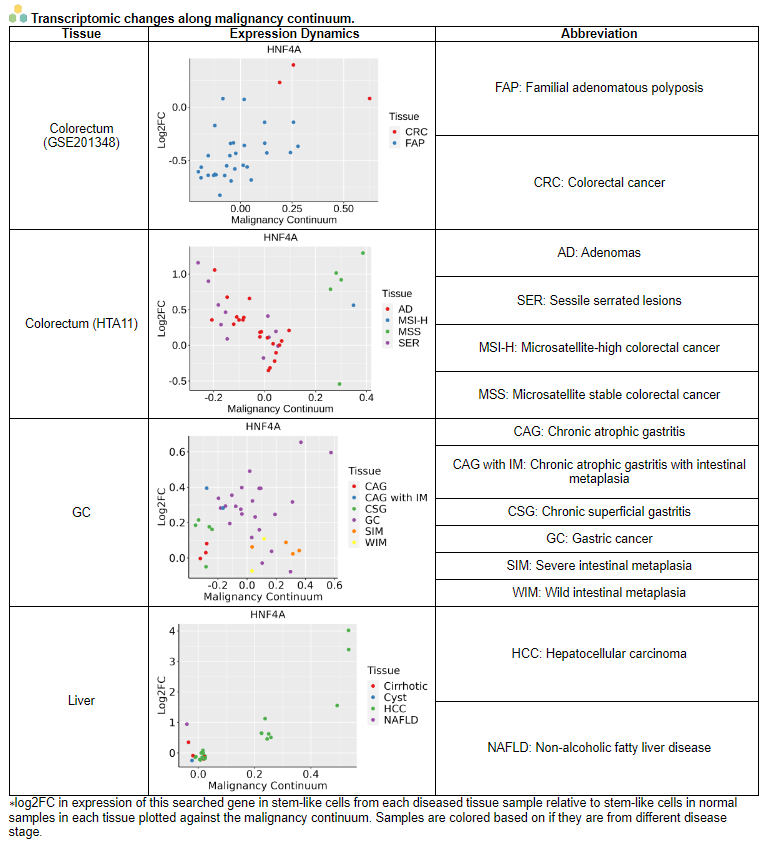
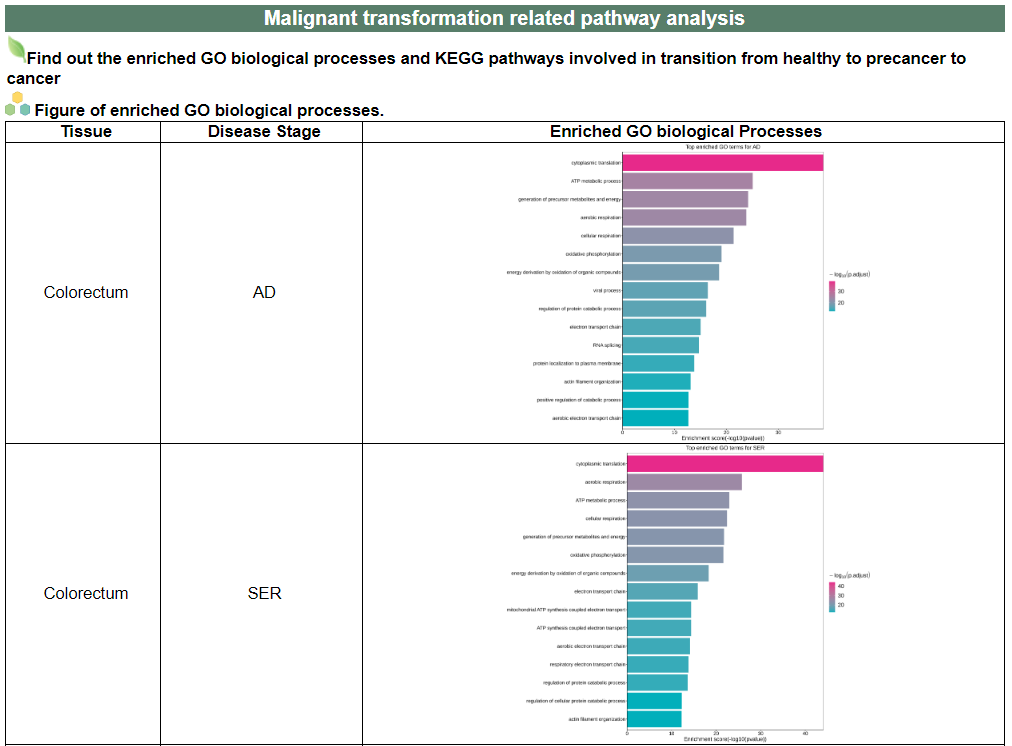
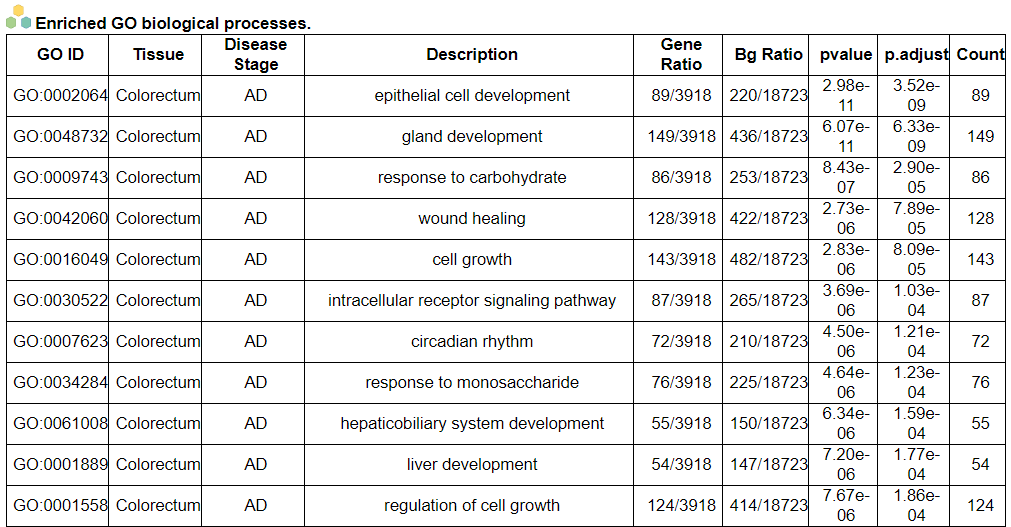
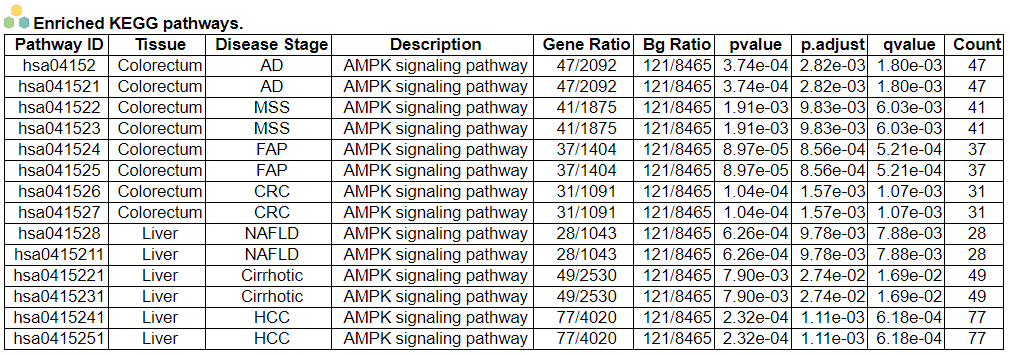
 iii. 'Malignant transformation related pathway analysis' category
iii. 'Malignant transformation related pathway analysis' categoryThis image category provides the result of malignant transformation related pathway analysis.
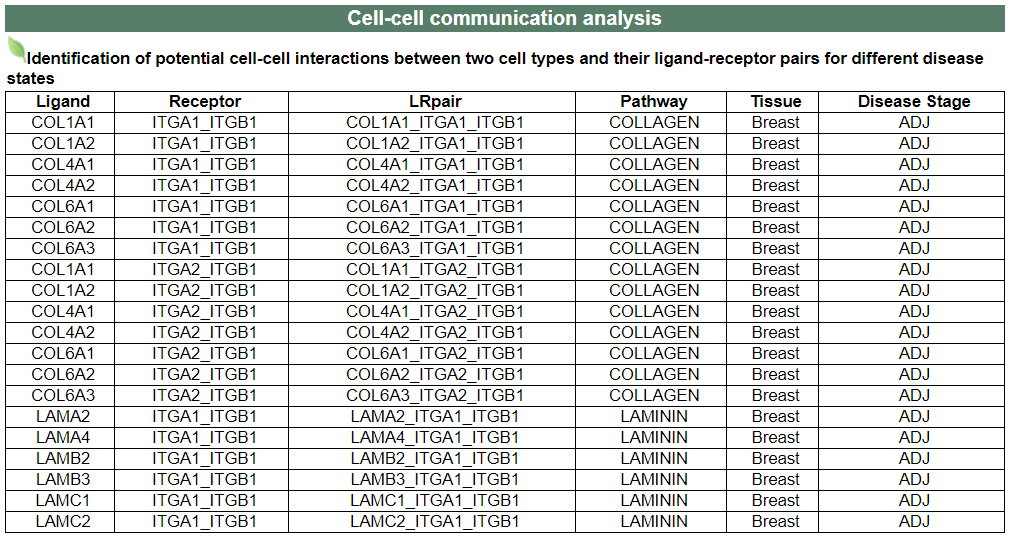
 iv. 'Cell-cell communication analysis' category
iv. 'Cell-cell communication analysis' categoryThis image category provides the result of cell-cell communication analysis.
Note: There is no result of HNF4A in this category. Here is the result of ITGB1 as an example.
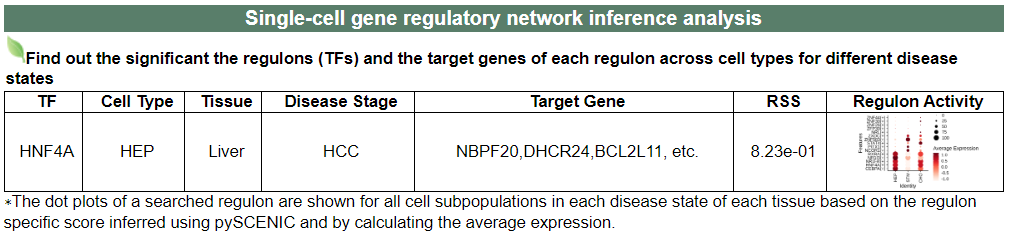
 v. 'Single-cell gene regulatory network inference analysis' category
v. 'Single-cell gene regulatory network inference analysis' categoryThis image category provides the result of single-cell gene regulatory network inference analysis.
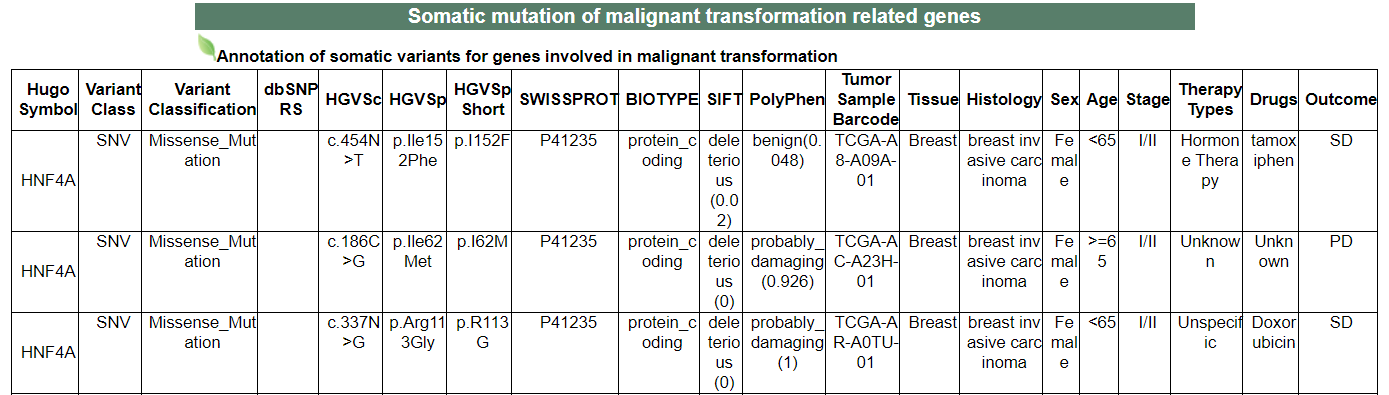
 vi. 'Somatic mutation of malignant transformation related genes' category
vi. 'Somatic mutation of malignant transformation related genes' categoryThis image category provides the somatic mutation of malignant transformation related genes.
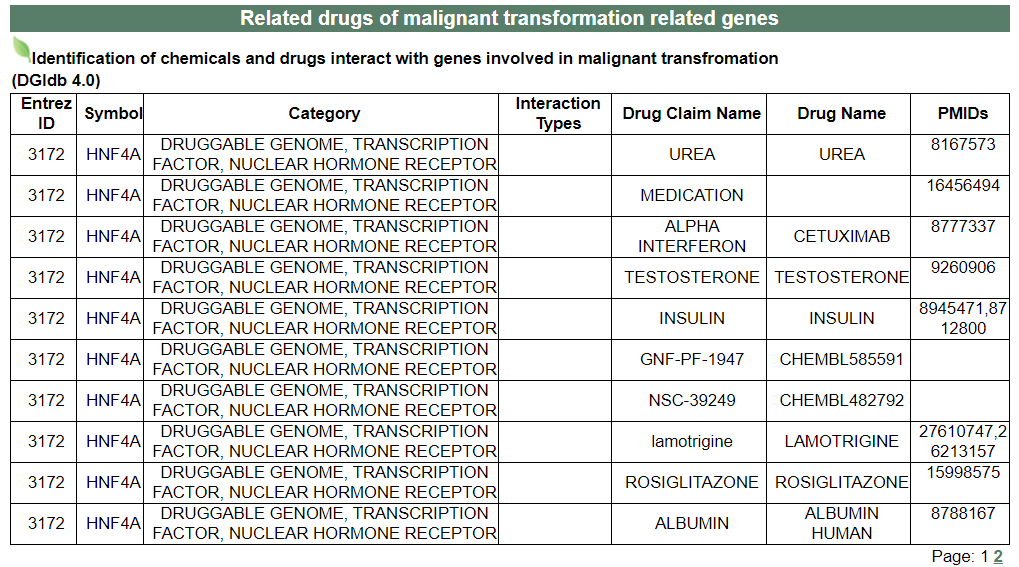
 vii. 'Related drugs of malignant transformation related genes' category
vii. 'Related drugs of malignant transformation related genes' categoryThis image category provides the related drugs of malignant transformation related genes.
Select your interested tissue.

This image shows the tissue annotation of PCTanno for your query with links to their corresponding annotation parts.

 i. 'Tissue samples summary' category
i. 'Tissue samples summary' categoryThis image category provides the tissue samples summary.
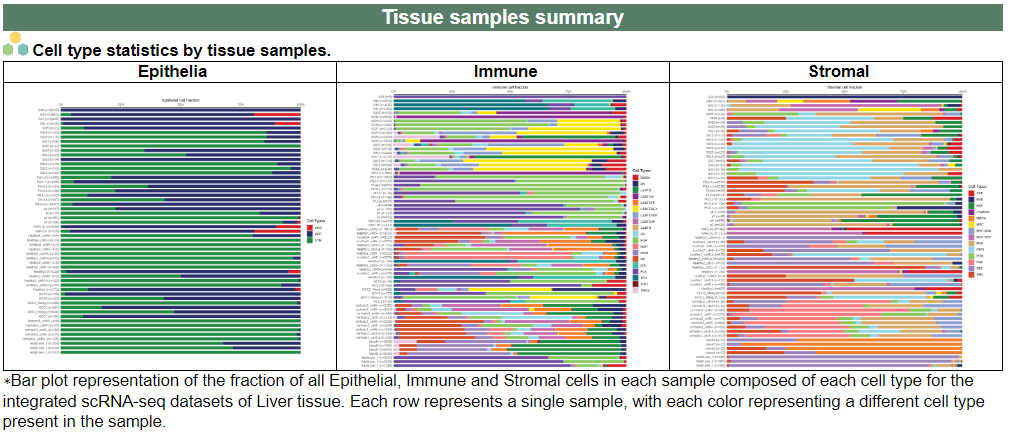
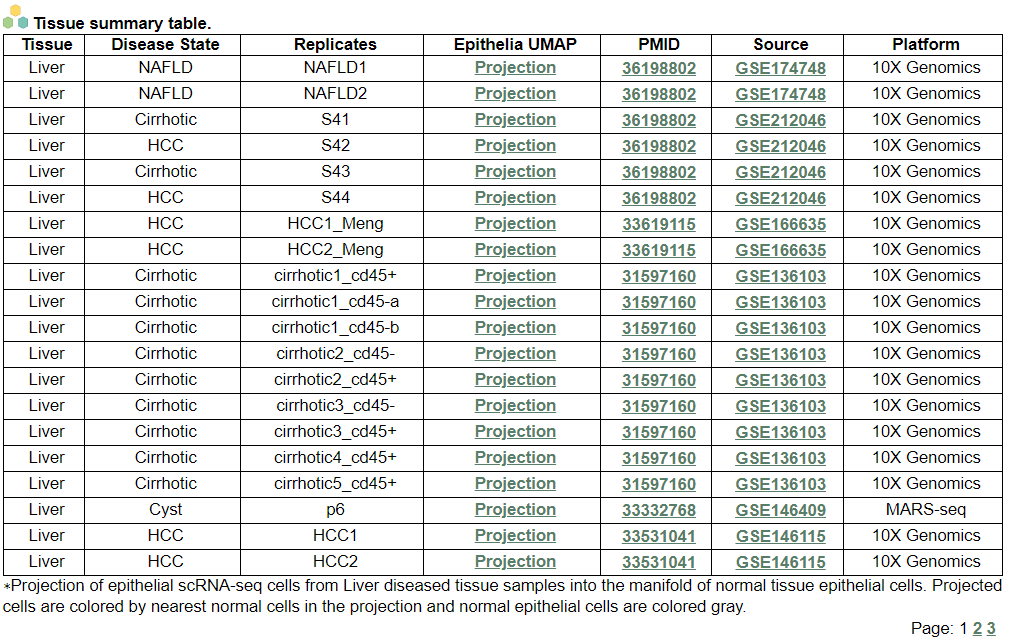
 ii. 'Annotation details for different cell compartments' category
ii. 'Annotation details for different cell compartments' categoryThis image category provides the annotation details for different cell compartments.
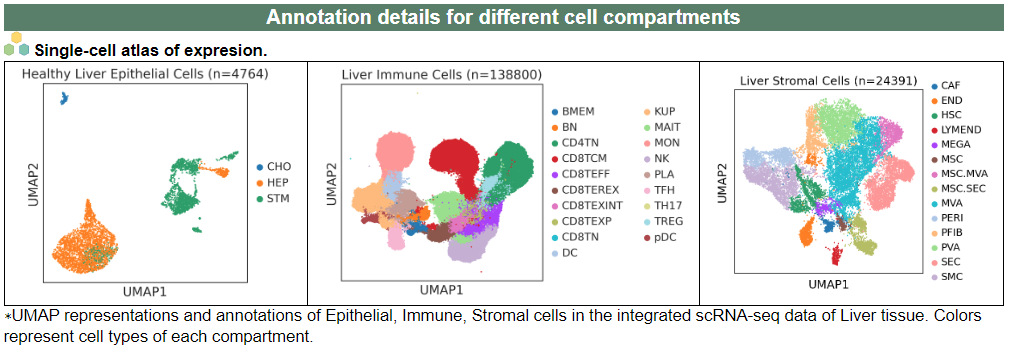
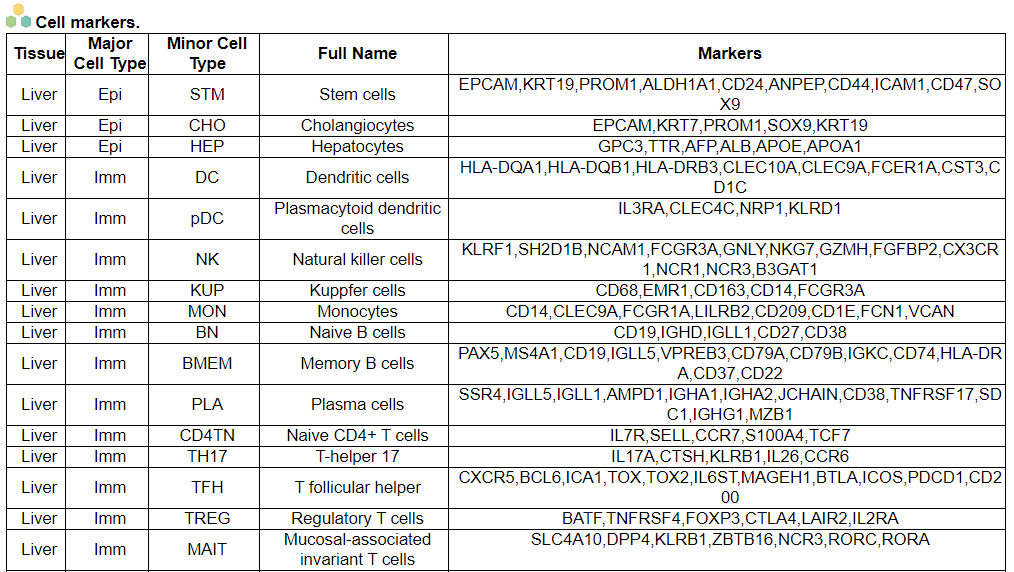
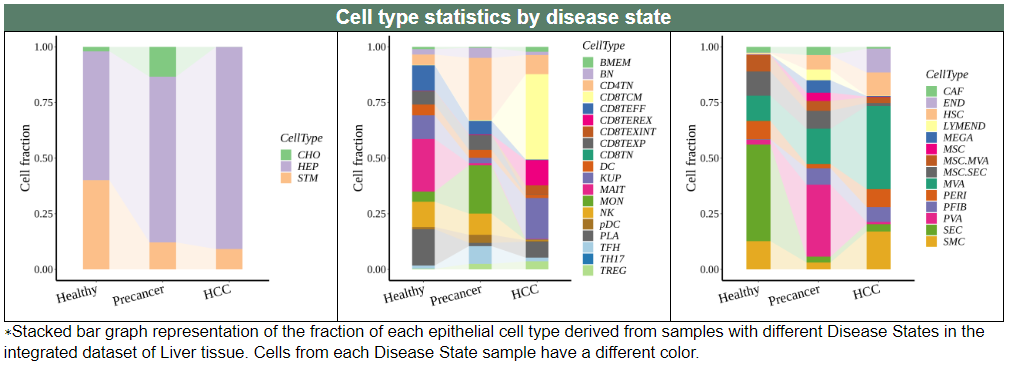
 iii. 'Cell type statistics by disease state' category
iii. 'Cell type statistics by disease state' categoryThis image category provides the cell type statistics by disease state.
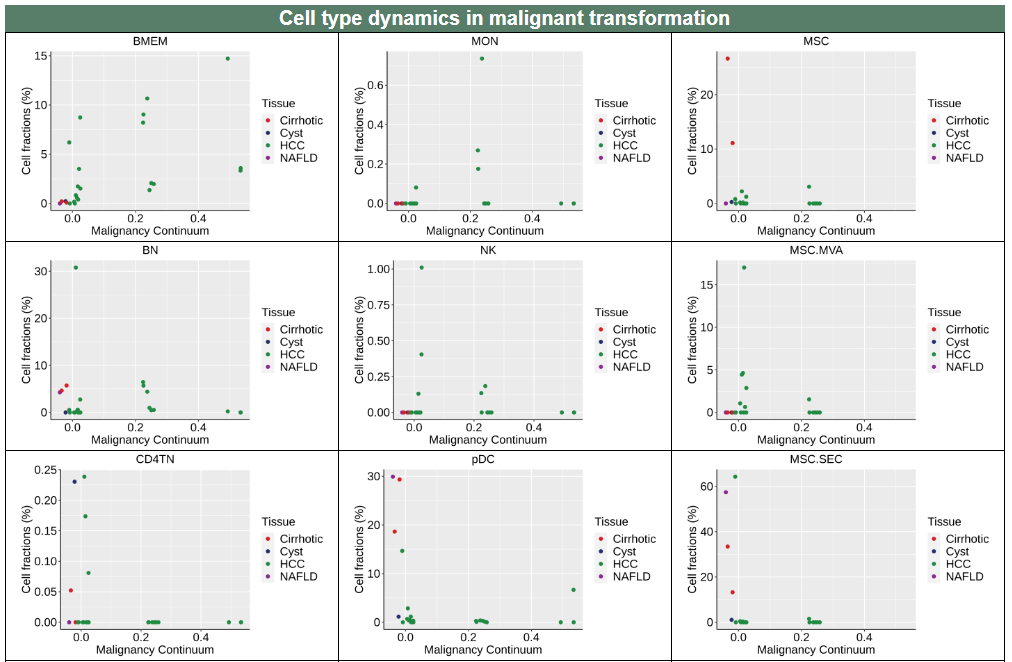
 iv. 'Cell type dynamics in malignant transformation' category
iv. 'Cell type dynamics in malignant transformation' categoryThis image category provides the cell type dynamics in malignant transformation.
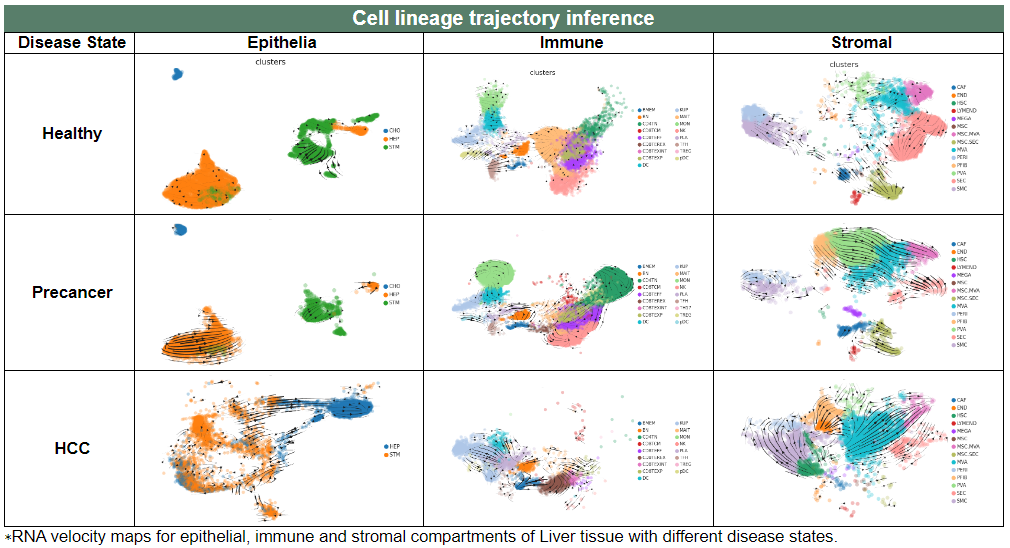
 v. 'Cell lineage trajectory inference' category
v. 'Cell lineage trajectory inference' categoryThis image category provides the result cell lineage trajectory inference.
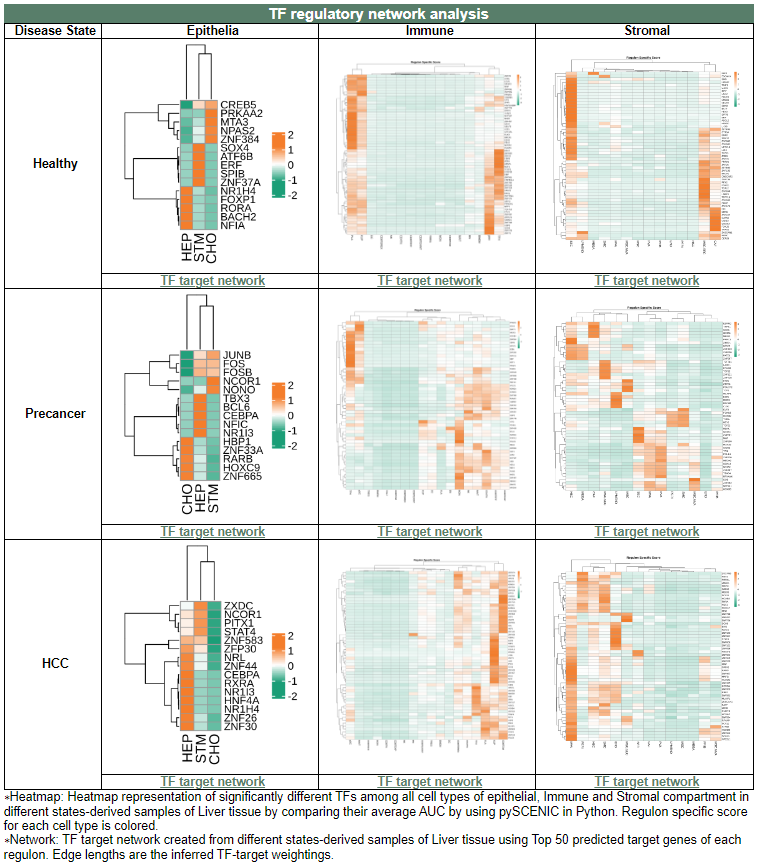
 vi. 'TF regulatory network analysis' category
vi. 'TF regulatory network analysis' categoryThis image category provides the result of TF regulatory network analysis.
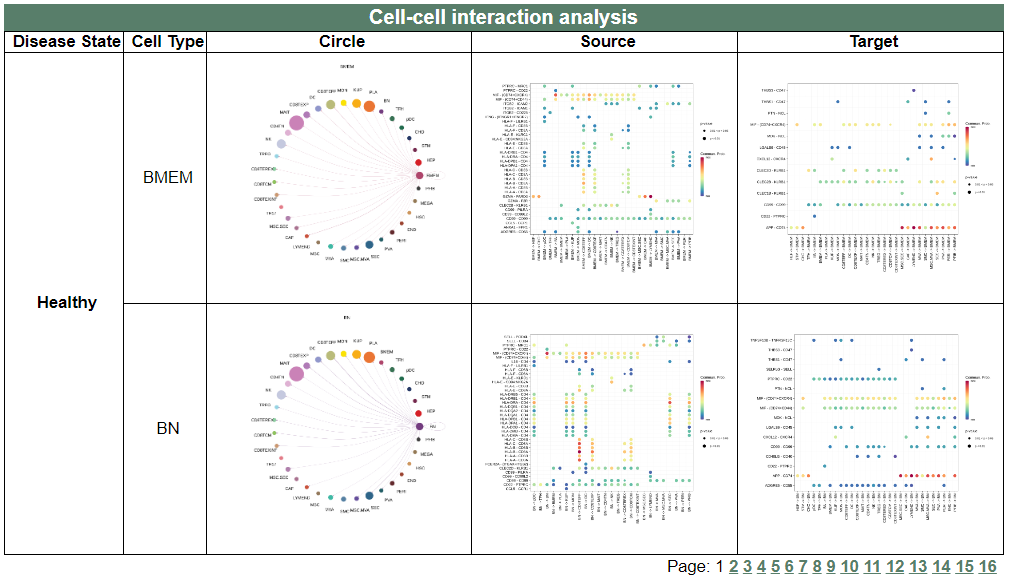
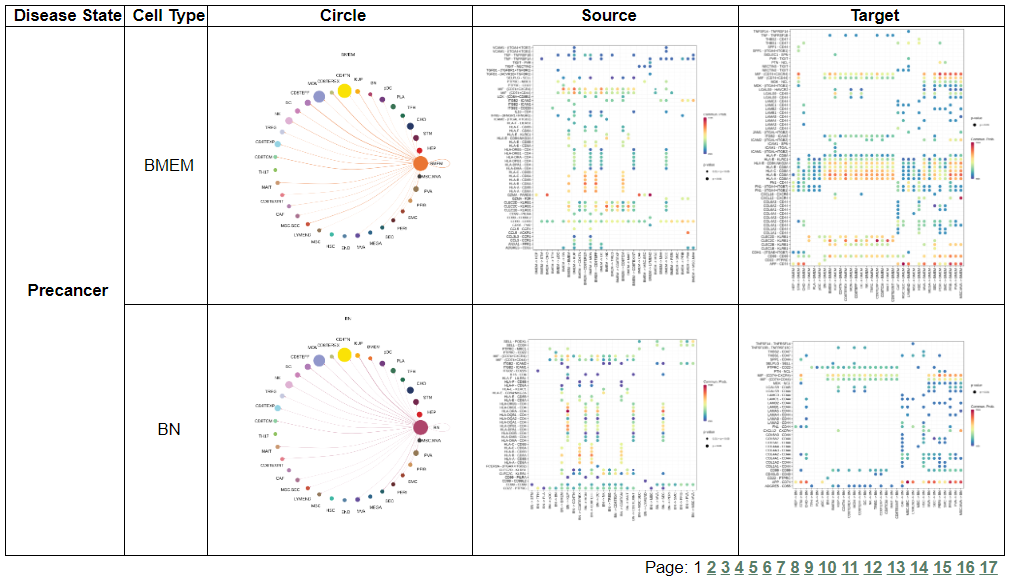
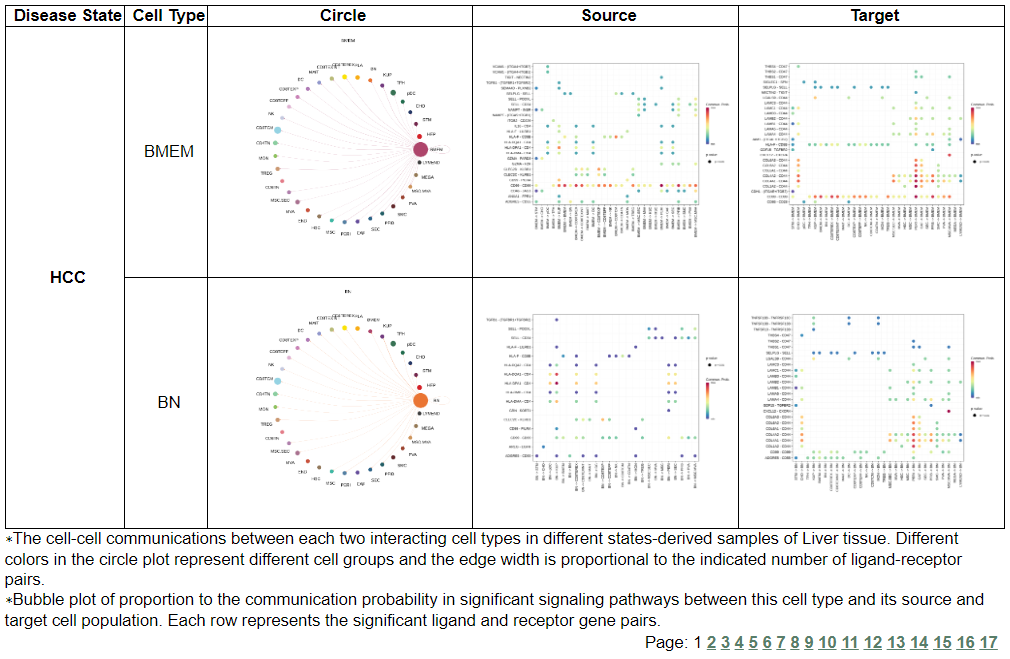
 vii. 'Cell-cell interaction analysis' category
vii. 'Cell-cell interaction analysis' categoryThis image category provides the result of cell-cell interaction analysis.
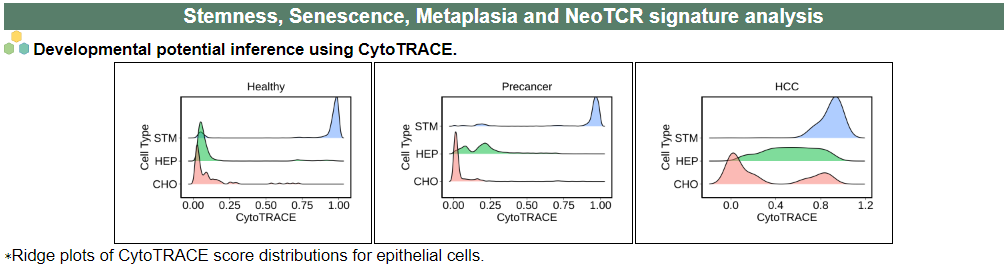
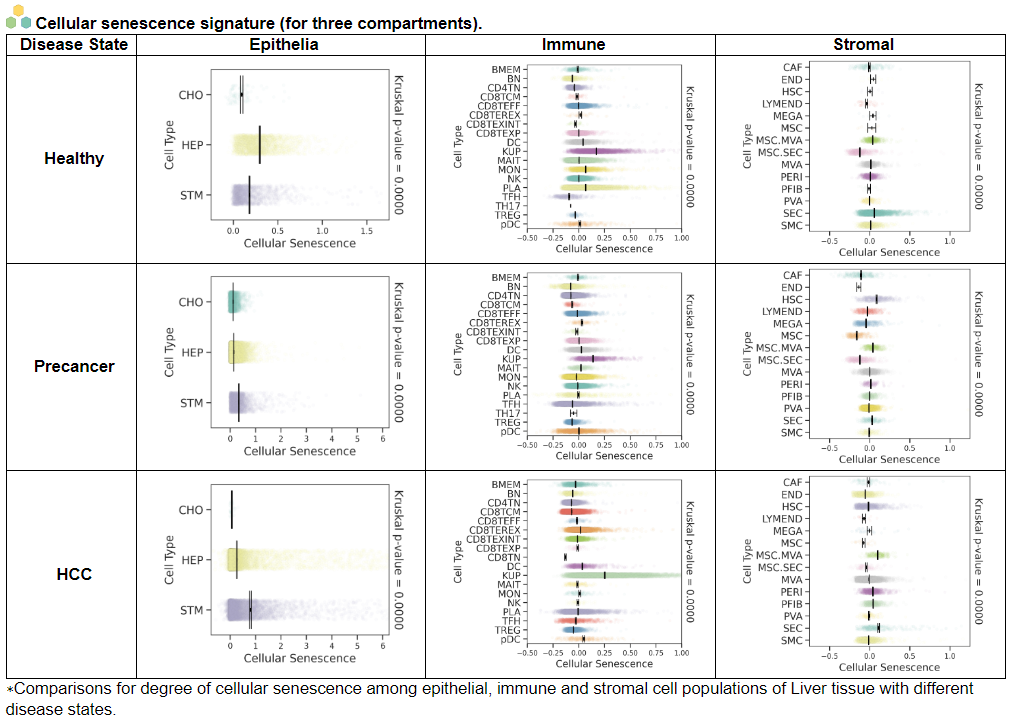
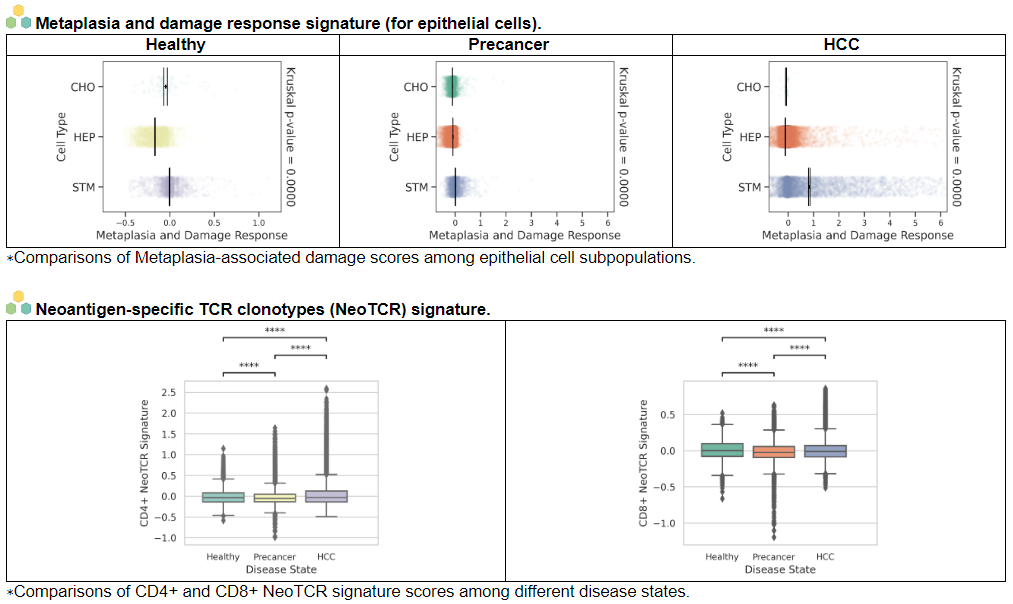
 viii. 'Stemness, Senescence, Metaplasia and NeoTCR signature analysis' category
viii. 'Stemness, Senescence, Metaplasia and NeoTCR signature analysis' categoryThis image category provides the result of Stemness, Senescence, Metaplasia and NeoTCR signature analysis.
Please go to download page and contact page.
