| GO ID | Tissue | Disease Stage | Description | Gene Ratio | Bg Ratio | pvalue | p.adjust | Count |
| GO:0006979113 | Thyroid | PTC | response to oxidative stress | 234/5968 | 446/18723 | 6.97e-20 | 9.77e-18 | 234 |
| GO:0062197113 | Thyroid | PTC | cellular response to chemical stress | 180/5968 | 337/18723 | 1.36e-16 | 1.16e-14 | 180 |
| GO:0032956113 | Thyroid | PTC | regulation of actin cytoskeleton organization | 182/5968 | 358/18723 | 4.64e-14 | 2.79e-12 | 182 |
| GO:0032970113 | Thyroid | PTC | regulation of actin filament-based process | 197/5968 | 397/18723 | 8.91e-14 | 4.89e-12 | 197 |
| GO:0034599113 | Thyroid | PTC | cellular response to oxidative stress | 151/5968 | 288/18723 | 2.82e-13 | 1.43e-11 | 151 |
| GO:0000302113 | Thyroid | PTC | response to reactive oxygen species | 121/5968 | 222/18723 | 2.10e-12 | 9.26e-11 | 121 |
| GO:0001701111 | Thyroid | PTC | in utero embryonic development | 175/5968 | 367/18723 | 1.40e-10 | 4.73e-09 | 175 |
| GO:0042060112 | Thyroid | PTC | wound healing | 190/5968 | 422/18723 | 7.72e-09 | 1.92e-07 | 190 |
| GO:0048732113 | Thyroid | PTC | gland development | 193/5968 | 436/18723 | 2.88e-08 | 6.42e-07 | 193 |
| GO:0034614112 | Thyroid | PTC | cellular response to reactive oxygen species | 82/5968 | 155/18723 | 4.32e-08 | 9.27e-07 | 82 |
| GO:004677716 | Thyroid | PTC | protein autophosphorylation | 110/5968 | 227/18723 | 1.25e-07 | 2.44e-06 | 110 |
| GO:003153220 | Thyroid | PTC | actin cytoskeleton reorganization | 60/5968 | 107/18723 | 1.91e-07 | 3.51e-06 | 60 |
| GO:0033674111 | Thyroid | PTC | positive regulation of kinase activity | 201/5968 | 467/18723 | 1.95e-07 | 3.54e-06 | 201 |
| GO:000182215 | Thyroid | PTC | kidney development | 133/5968 | 293/18723 | 7.57e-07 | 1.18e-05 | 133 |
| GO:000165517 | Thyroid | PTC | urogenital system development | 149/5968 | 338/18723 | 1.41e-06 | 2.01e-05 | 149 |
| GO:007200114 | Thyroid | PTC | renal system development | 135/5968 | 302/18723 | 1.76e-06 | 2.43e-05 | 135 |
| GO:000110119 | Thyroid | PTC | response to acid chemical | 69/5968 | 135/18723 | 2.55e-06 | 3.34e-05 | 69 |
| GO:0048511111 | Thyroid | PTC | rhythmic process | 131/5968 | 298/18723 | 7.06e-06 | 8.33e-05 | 131 |
| GO:00480089 | Thyroid | PTC | platelet-derived growth factor receptor signaling pathway | 34/5968 | 56/18723 | 8.20e-06 | 9.42e-05 | 34 |
| GO:0031032110 | Thyroid | PTC | actomyosin structure organization | 91/5968 | 196/18723 | 1.35e-05 | 1.45e-04 | 91 |
| Hugo Symbol | Variant Class | Variant Classification | dbSNP RS | HGVSc | HGVSp | HGVSp Short | SWISSPROT | BIOTYPE | SIFT | PolyPhen | Tumor Sample Barcode | Tissue | Histology | Sex | Age | Stage | Therapy Types | Drugs | Outcome |
| PDGFRA | SNV | Missense_Mutation | | c.2942G>C | p.Arg981Pro | p.R981P | P16234 | protein_coding | deleterious(0.03) | possibly_damaging(0.862) | TCGA-A2-A0T0-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Chemotherapy | taxotere | SD |
| PDGFRA | SNV | Missense_Mutation | | c.2953N>A | p.Asp985Asn | p.D985N | P16234 | protein_coding | tolerated(0.27) | benign(0.318) | TCGA-AC-A23H-01 | Breast | breast invasive carcinoma | Female | >=65 | I/II | Unknown | Unknown | PD |
| PDGFRA | SNV | Missense_Mutation | | c.1711N>A | p.Glu571Lys | p.E571K | P16234 | protein_coding | deleterious(0) | probably_damaging(0.989) | TCGA-AO-A12D-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Chemotherapy | cyclophosphamide | SD |
| PDGFRA | SNV | Missense_Mutation | rs763325080 | c.2465N>A | p.Arg822His | p.R822H | P16234 | protein_coding | deleterious(0) | probably_damaging(0.996) | TCGA-AR-A1AV-01 | Breast | breast invasive carcinoma | Male | >=65 | I/II | Chemotherapy | cytoxan | SD |
| PDGFRA | SNV | Missense_Mutation | rs767697835 | c.3223N>A | p.Asp1075Asn | p.D1075N | P16234 | protein_coding | deleterious_low_confidence(0.01) | possibly_damaging(0.631) | TCGA-D8-A1XQ-01 | Breast | breast invasive carcinoma | Female | >=65 | I/II | Unknown | Unknown | SD |
| PDGFRA | SNV | Missense_Mutation | | c.1666G>A | p.Glu556Lys | p.E556K | P16234 | protein_coding | deleterious(0) | probably_damaging(0.993) | TCGA-D8-A27G-01 | Breast | breast invasive carcinoma | Female | >=65 | I/II | Unknown | Unknown | SD |
| PDGFRA | insertion | Frame_Shift_Ins | novel | c.905_906insATCTCCATCTCTTAGAATCTTTCTCCACCATCTCTCCTTTCTAG | p.Met302IlefsTer51 | p.M302Ifs*51 | P16234 | protein_coding | | | TCGA-A2-A0CX-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Chemotherapy | adriamycin | SD |
| PDGFRA | insertion | Nonsense_Mutation | novel | c.2875_2876insCCATGTTCAGCTAATTTTTGTATTTTT | p.Lys959delinsThrMetPheSerTerPheLeuTyrPheTer | p.K959delinsTMFS*FLYF* | P16234 | protein_coding | | | TCGA-A7-A0CJ-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Chemotherapy | cytoxan | SD |
| PDGFRA | insertion | Frame_Shift_Ins | novel | c.2035_2036insTG | p.Tyr679LeufsTer17 | p.Y679Lfs*17 | P16234 | protein_coding | | | TCGA-A8-A06O-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Hormone Therapy | letrozole | SD |
| PDGFRA | insertion | Nonsense_Mutation | novel | c.2036_2037insGAACAGGAAGT | p.Tyr679Ter | p.Y679* | P16234 | protein_coding | | | TCGA-A8-A06O-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Hormone Therapy | letrozole | SD |
| Entrez ID | Symbol | Category | Interaction Types | Drug Claim Name | Drug Name | PMIDs |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | | IMATINIB | IMATINIB | 15928335,15685537,22718859,16638875,15146165,12949711,26130666,25157968,24132921,14645423,16954519,18794084,22745105 |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | | DASATINIB | DASATINIB | 15928335,15685537,22718859,16638875,15146165,12949711,26130666,25157968,14645423,16954519,18794084,22745105 |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | | VANDETANIB | VANDETANIB | |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | inhibitor | 363894199 | | |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | | CP-868596 | CRENOLANIB | |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | inhibitor | 249565576 | ENMD-2076 | |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | | LINIFANIB | LINIFANIB | |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | antibody | OLARATUMAB | OLARATUMAB | |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | inhibitor | 178102559 | NINTEDANIB | |
| 5156 | PDGFRA | TYROSINE KINASE, KINASE, CLINICALLY ACTIONABLE, DRUGGABLE GENOME, EXTERNAL SIDE OF PLASMA MEMBRANE, DRUG RESISTANCE | | CYC-116 | CYC-116 | |







 Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells
Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells
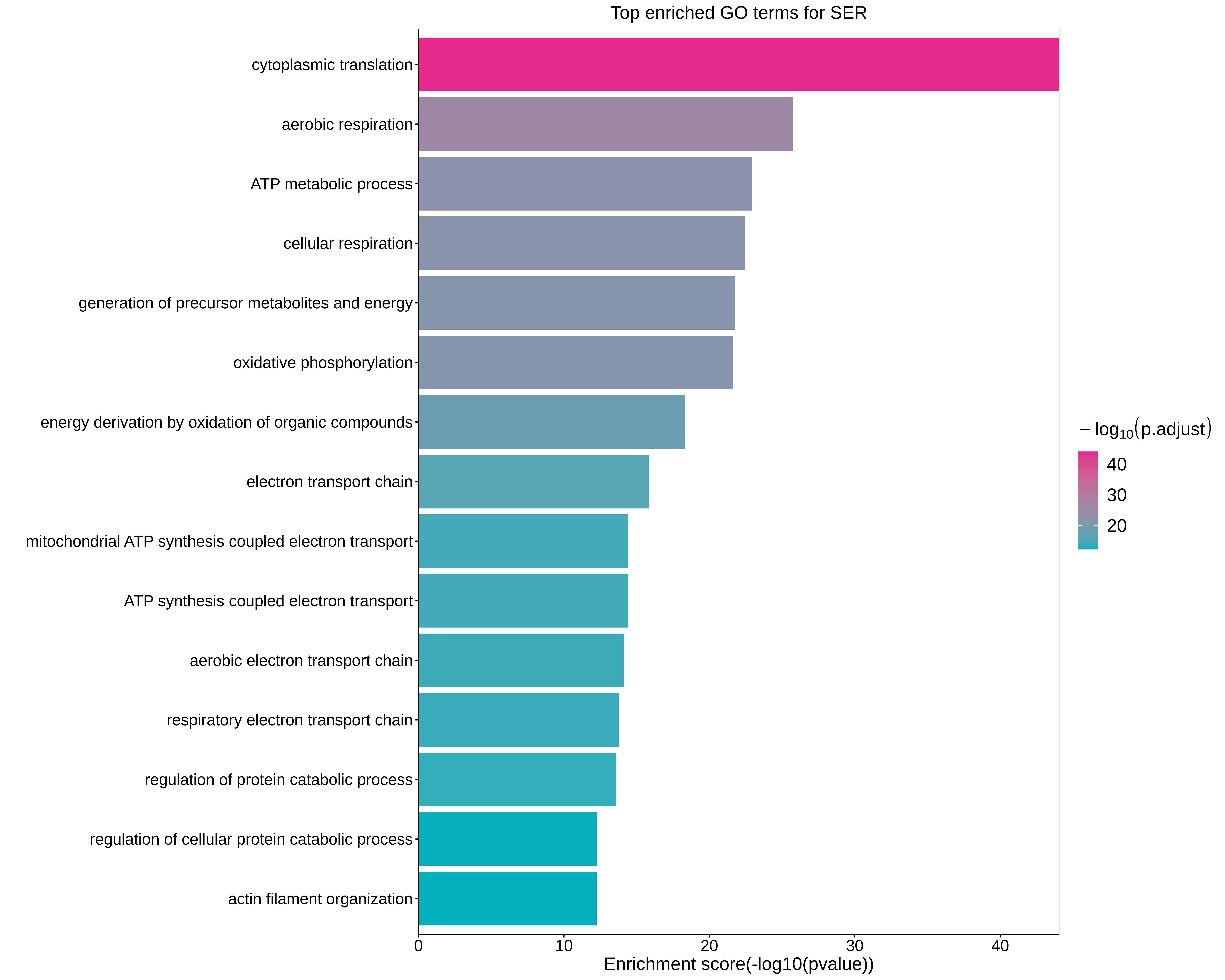
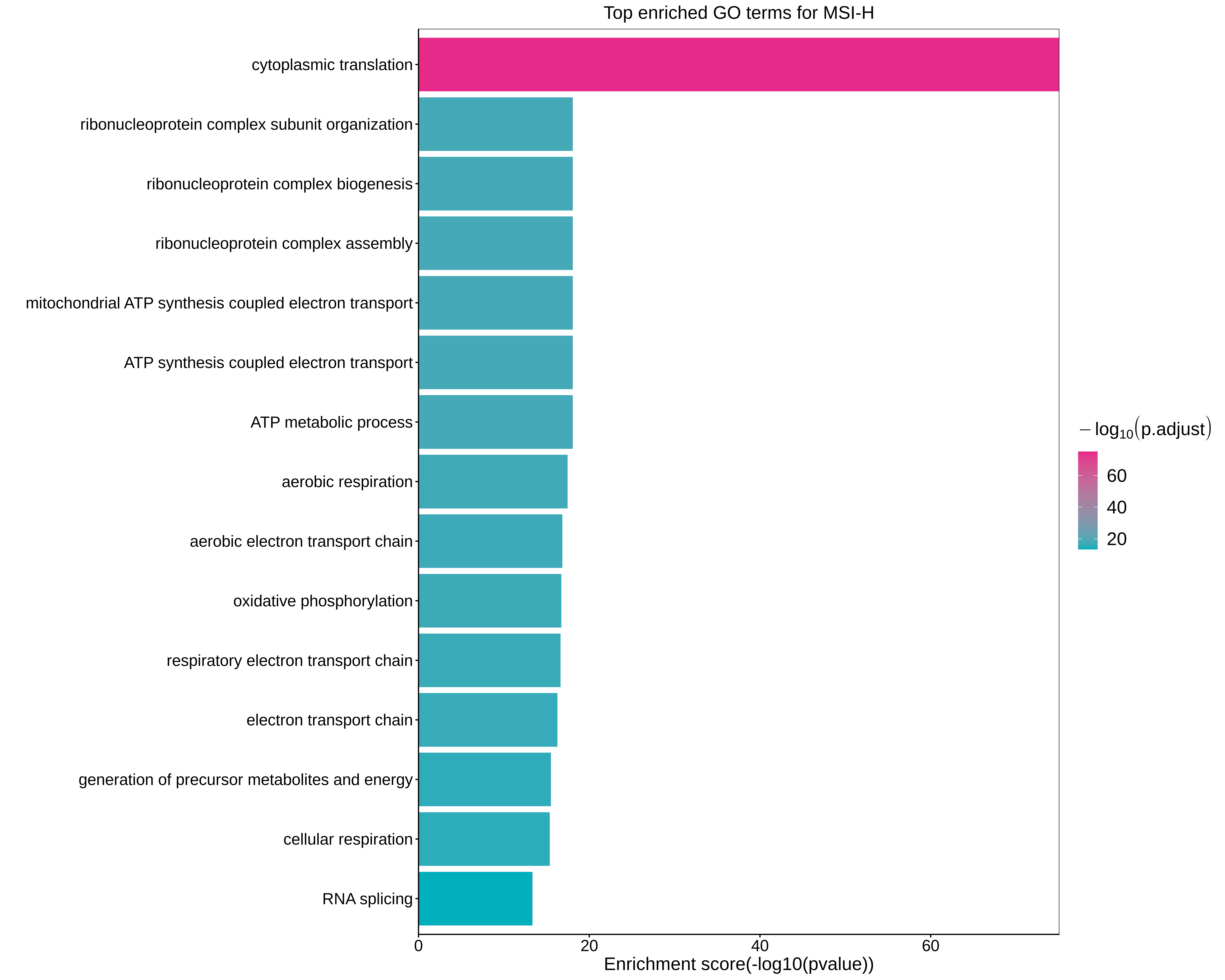
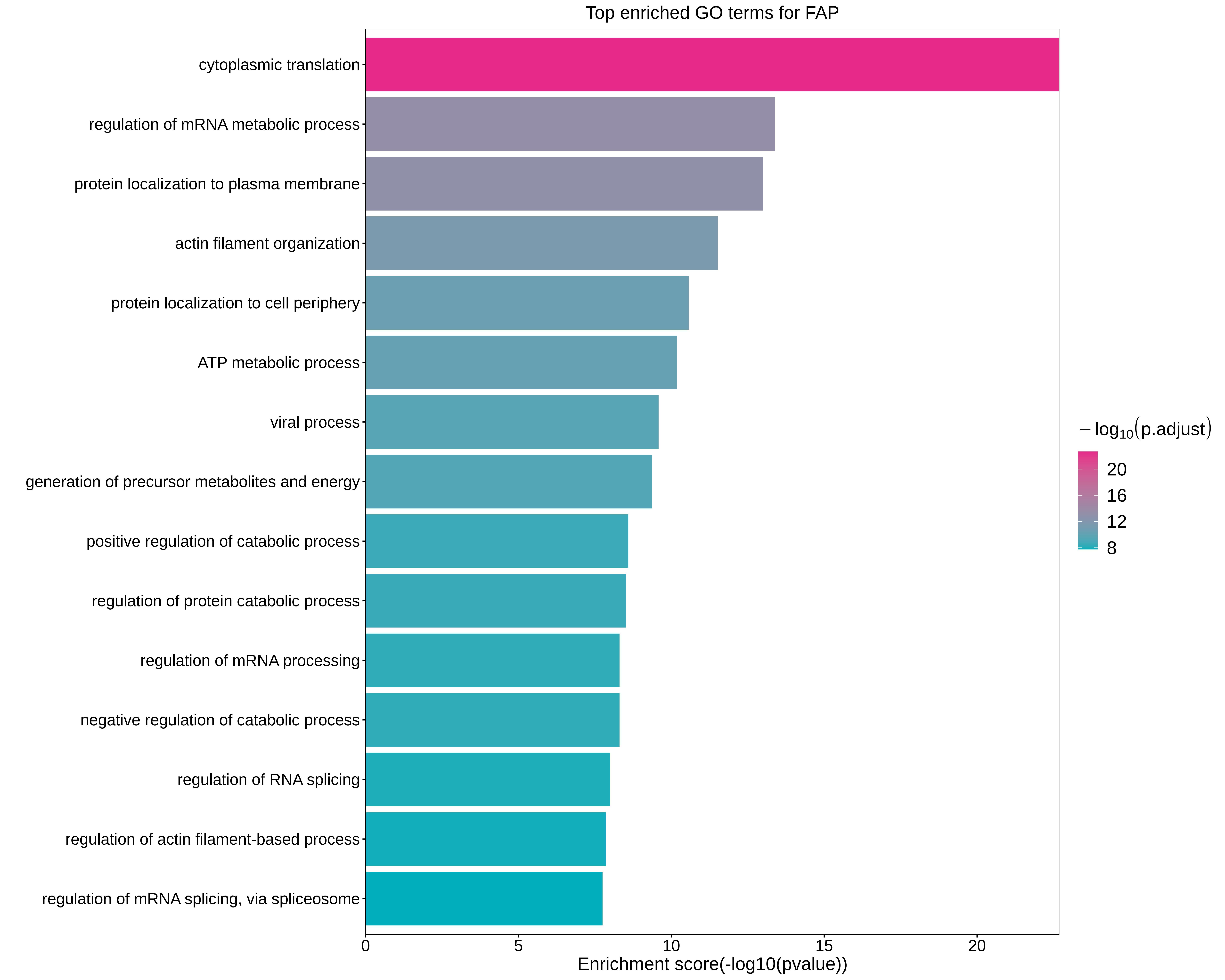
 Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer
Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer

 Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states
Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states
Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states Annotation of somatic variants for genes involved in malignant transformation
Annotation of somatic variants for genes involved in malignant transformation Identification of chemicals and drugs interact with genes involved in malignant transfromation
Identification of chemicals and drugs interact with genes involved in malignant transfromation