| Tissue | Expression Dynamics | Abbreviation |
| Esophagus |  | ESCC: Esophageal squamous cell carcinoma |
| HGIN: High-grade intraepithelial neoplasias |
| LGIN: Low-grade intraepithelial neoplasias |
| Liver | 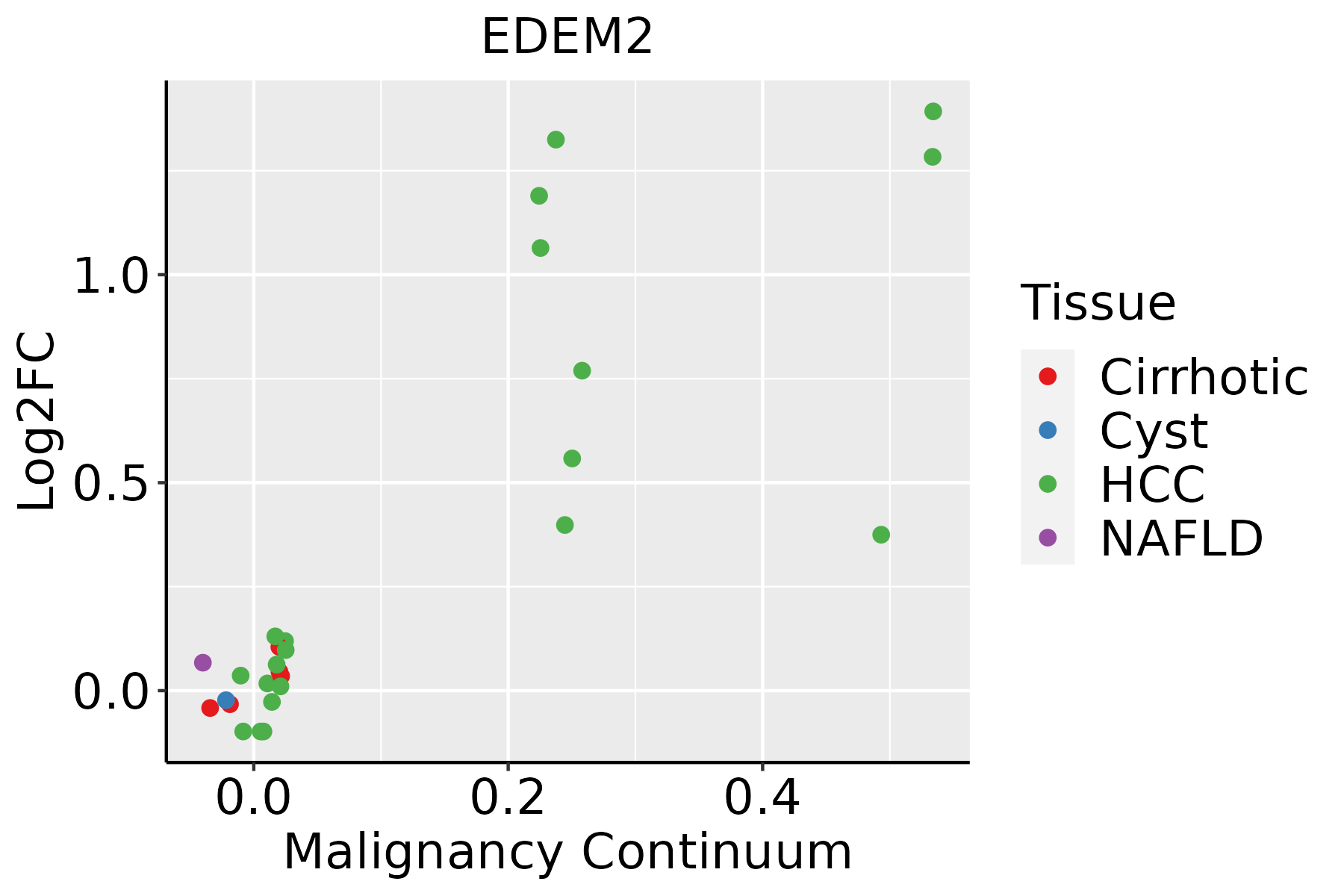 | HCC: Hepatocellular carcinoma |
| NAFLD: Non-alcoholic fatty liver disease |
| Oral Cavity | 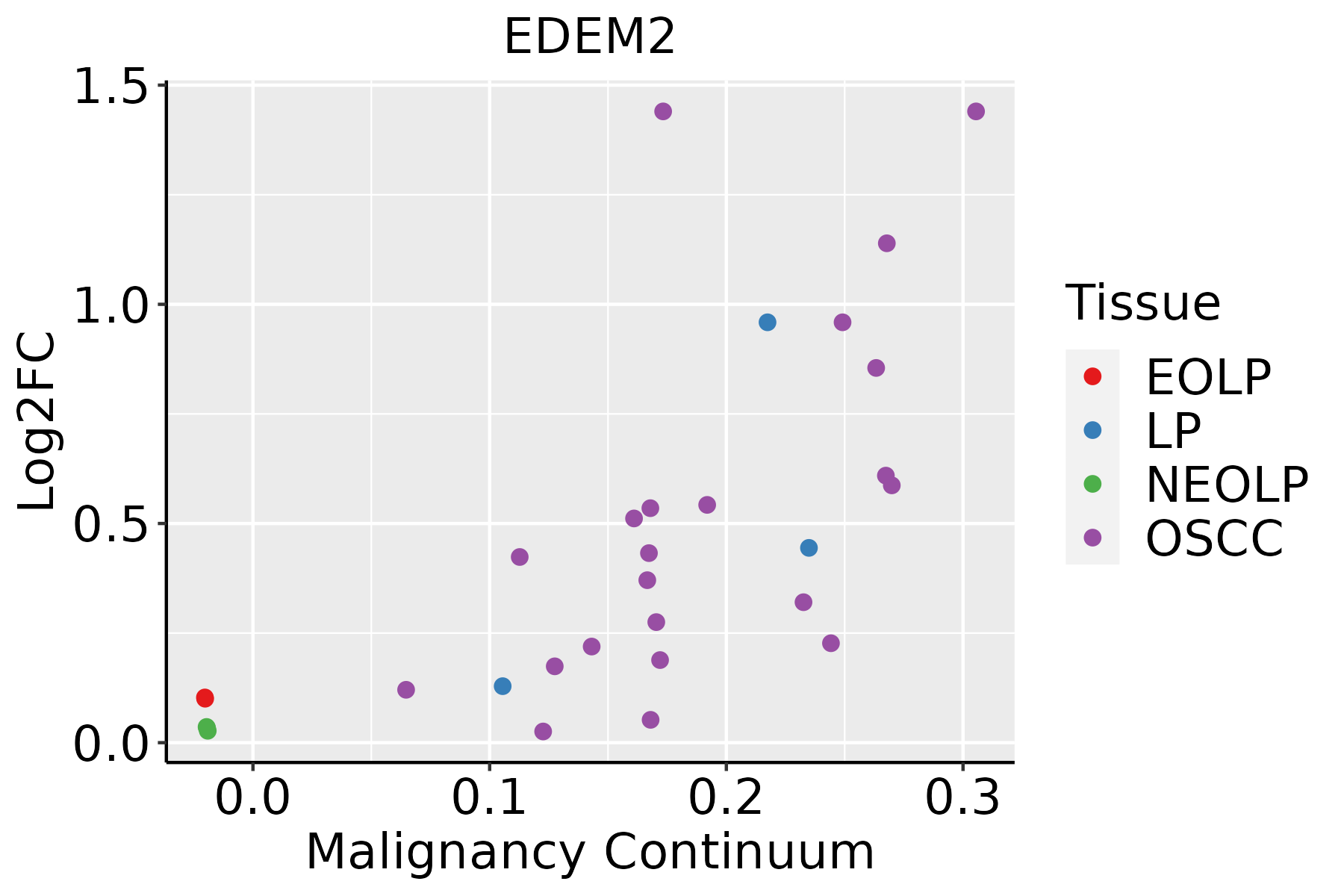 | EOLP: Erosive Oral lichen planus |
| LP: leukoplakia |
| NEOLP: Non-erosive oral lichen planus |
| OSCC: Oral squamous cell carcinoma |
| Thyroid | 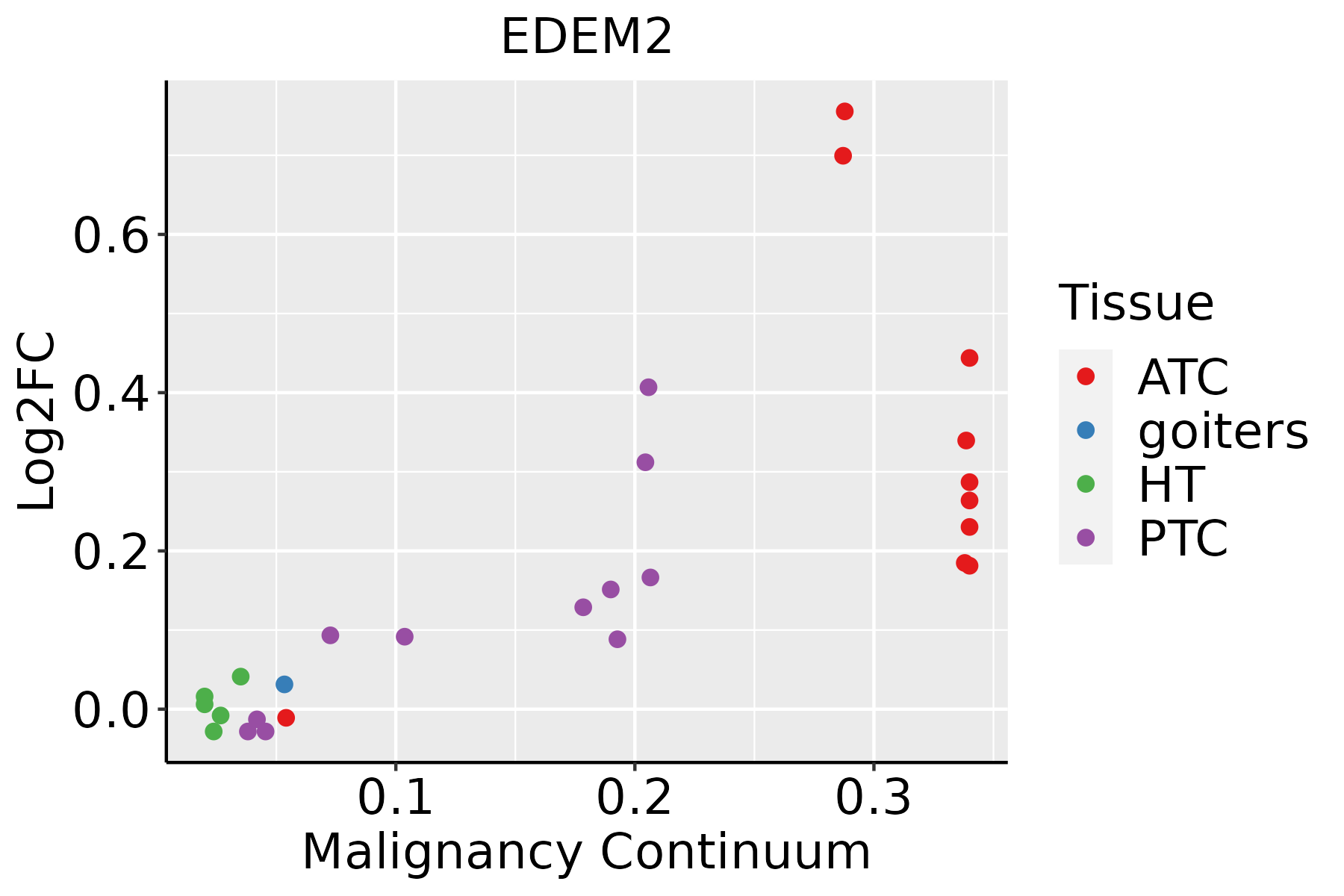 | ATC: Anaplastic thyroid cancer |
| HT: Hashimoto's thyroiditis |
| PTC: Papillary thyroid cancer |
| GO ID | Tissue | Disease Stage | Description | Gene Ratio | Bg Ratio | pvalue | p.adjust | Count |
| GO:001603234 | Thyroid | ATC | viral process | 241/6293 | 415/18723 | 5.50e-25 | 2.04e-22 | 241 |
| GO:003238632 | Thyroid | ATC | regulation of intracellular transport | 203/6293 | 337/18723 | 6.16e-24 | 1.85e-21 | 203 |
| GO:003315726 | Thyroid | ATC | regulation of intracellular protein transport | 147/6293 | 229/18723 | 2.01e-21 | 4.38e-19 | 147 |
| GO:003238825 | Thyroid | ATC | positive regulation of intracellular transport | 131/6293 | 202/18723 | 8.20e-20 | 1.24e-17 | 131 |
| GO:009031624 | Thyroid | ATC | positive regulation of intracellular protein transport | 107/6293 | 160/18723 | 6.82e-18 | 7.84e-16 | 107 |
| GO:190495128 | Thyroid | ATC | positive regulation of establishment of protein localization | 177/6293 | 319/18723 | 5.43e-16 | 4.29e-14 | 177 |
| GO:003596633 | Thyroid | ATC | response to topologically incorrect protein | 102/6293 | 159/18723 | 2.87e-15 | 1.99e-13 | 102 |
| GO:005122234 | Thyroid | ATC | positive regulation of protein transport | 165/6293 | 303/18723 | 4.94e-14 | 2.69e-12 | 165 |
| GO:000698633 | Thyroid | ATC | response to unfolded protein | 88/6293 | 137/18723 | 2.04e-13 | 9.91e-12 | 88 |
| GO:003650322 | Thyroid | ATC | ERAD pathway | 72/6293 | 107/18723 | 1.05e-12 | 4.41e-11 | 72 |
| GO:003043322 | Thyroid | ATC | ubiquitin-dependent ERAD pathway | 60/6293 | 85/18723 | 3.38e-12 | 1.38e-10 | 60 |
| GO:001908023 | Thyroid | ATC | viral gene expression | 64/6293 | 94/18723 | 8.44e-12 | 3.16e-10 | 64 |
| GO:003097015 | Thyroid | ATC | retrograde protein transport, ER to cytosol | 23/6293 | 29/18723 | 5.84e-07 | 8.19e-06 | 23 |
| GO:190351315 | Thyroid | ATC | endoplasmic reticulum to cytosol transport | 23/6293 | 29/18723 | 5.84e-07 | 8.19e-06 | 23 |
| GO:003252715 | Thyroid | ATC | protein exit from endoplasmic reticulum | 32/6293 | 48/18723 | 2.88e-06 | 3.30e-05 | 32 |
| GO:001908213 | Thyroid | ATC | viral protein processing | 18/6293 | 29/18723 | 1.58e-03 | 7.98e-03 | 18 |
| GO:007086113 | Thyroid | ATC | regulation of protein exit from endoplasmic reticulum | 17/6293 | 27/18723 | 1.70e-03 | 8.47e-03 | 17 |
| GO:190415212 | Thyroid | ATC | regulation of retrograde protein transport, ER to cytosol | 10/6293 | 14/18723 | 4.31e-03 | 1.80e-02 | 10 |
| GO:000910011 | Thyroid | ATC | glycoprotein metabolic process | 153/6293 | 387/18723 | 7.90e-03 | 3.06e-02 | 153 |
| GO:19011363 | Thyroid | ATC | carbohydrate derivative catabolic process | 73/6293 | 172/18723 | 9.43e-03 | 3.47e-02 | 73 |
| Pathway ID | Tissue | Disease Stage | Description | Gene Ratio | Bg Ratio | pvalue | p.adjust | qvalue | Count |
| hsa04141211 | Esophagus | ESCC | Protein processing in endoplasmic reticulum | 147/4205 | 174/8465 | 3.29e-22 | 1.10e-19 | 5.64e-20 | 147 |
| hsa04141310 | Esophagus | ESCC | Protein processing in endoplasmic reticulum | 147/4205 | 174/8465 | 3.29e-22 | 1.10e-19 | 5.64e-20 | 147 |
| hsa0414122 | Liver | HCC | Protein processing in endoplasmic reticulum | 146/4020 | 174/8465 | 7.34e-24 | 2.46e-21 | 1.37e-21 | 146 |
| hsa0414132 | Liver | HCC | Protein processing in endoplasmic reticulum | 146/4020 | 174/8465 | 7.34e-24 | 2.46e-21 | 1.37e-21 | 146 |
| hsa0414130 | Oral cavity | OSCC | Protein processing in endoplasmic reticulum | 143/3704 | 174/8465 | 6.82e-26 | 2.28e-23 | 1.16e-23 | 143 |
| hsa04141113 | Oral cavity | OSCC | Protein processing in endoplasmic reticulum | 143/3704 | 174/8465 | 6.82e-26 | 2.28e-23 | 1.16e-23 | 143 |
| hsa04141210 | Oral cavity | LP | Protein processing in endoplasmic reticulum | 113/2418 | 174/8465 | 8.74e-24 | 5.82e-22 | 3.76e-22 | 113 |
| hsa0414138 | Oral cavity | LP | Protein processing in endoplasmic reticulum | 113/2418 | 174/8465 | 8.74e-24 | 5.82e-22 | 3.76e-22 | 113 |
| hsa0414145 | Oral cavity | EOLP | Protein processing in endoplasmic reticulum | 70/1218 | 174/8465 | 2.84e-17 | 3.06e-15 | 1.81e-15 | 70 |
| hsa0414155 | Oral cavity | EOLP | Protein processing in endoplasmic reticulum | 70/1218 | 174/8465 | 2.84e-17 | 3.06e-15 | 1.81e-15 | 70 |
| Hugo Symbol | Variant Class | Variant Classification | dbSNP RS | HGVSc | HGVSp | HGVSp Short | SWISSPROT | BIOTYPE | SIFT | PolyPhen | Tumor Sample Barcode | Tissue | Histology | Sex | Age | Stage | Therapy Types | Drugs | Outcome |
| EDEM2 | SNV | Missense_Mutation | | c.1079C>A | p.Thr360Lys | p.T360K | Q9BV94 | protein_coding | deleterious(0.03) | possibly_damaging(0.725) | TCGA-A8-A075-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Chemotherapy | epirubicin | CR |
| EDEM2 | SNV | Missense_Mutation | rs140405715 | c.1132N>G | p.Met378Val | p.M378V | Q9BV94 | protein_coding | deleterious(0.04) | possibly_damaging(0.718) | TCGA-A8-A08S-01 | Breast | breast invasive carcinoma | Female | >=65 | I/II | Hormone Therapy | anastrozole | SD |
| EDEM2 | SNV | Missense_Mutation | novel | c.772N>A | p.Asp258Asn | p.D258N | Q9BV94 | protein_coding | deleterious(0) | probably_damaging(1) | TCGA-AC-A62V-01 | Breast | breast invasive carcinoma | Male | <65 | III/IV | Targeted Molecular therapy | denosumab | PD |
| EDEM2 | SNV | Missense_Mutation | rs754555510 | c.476N>A | p.Arg159Gln | p.R159Q | Q9BV94 | protein_coding | tolerated(0.29) | benign(0.145) | TCGA-AN-A0AK-01 | Breast | breast invasive carcinoma | Female | >=65 | I/II | Unknown | Unknown | SD |
| EDEM2 | SNV | Missense_Mutation | rs146309337 | c.1565N>T | p.Ser522Leu | p.S522L | Q9BV94 | protein_coding | tolerated(0.32) | benign(0) | TCGA-B6-A402-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Chemotherapy | cyclophosphamide | PD |
| EDEM2 | SNV | Missense_Mutation | | c.1706T>C | p.Leu569Ser | p.L569S | Q9BV94 | protein_coding | deleterious_low_confidence(0) | benign(0.075) | TCGA-D8-A1XQ-01 | Breast | breast invasive carcinoma | Female | >=65 | I/II | Unknown | Unknown | SD |
| EDEM2 | SNV | Missense_Mutation | novel | c.1042N>C | p.Gly348Arg | p.G348R | Q9BV94 | protein_coding | tolerated(0.06) | probably_damaging(0.964) | TCGA-GI-A2C9-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Unspecific | | SD |
| EDEM2 | insertion | In_Frame_Ins | novel | c.300_301insTCATTCTTATAC | p.Val100_Leu101insSerPheLeuTyr | p.V100_L101insSFLY | Q9BV94 | protein_coding | | | TCGA-B6-A0IA-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Unknown | Unknown | SD |
| EDEM2 | insertion | Nonsense_Mutation | novel | c.299_300insTTGTCATCTGTAAATGAAG | p.Leu101CysfsTer4 | p.L101Cfs*4 | Q9BV94 | protein_coding | | | TCGA-B6-A0IA-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Unknown | Unknown | SD |
| EDEM2 | insertion | Frame_Shift_Ins | novel | c.761_762insAGTGCCTGCAGAATCAGTTCTGAATTCAGAGTCACTAT | p.Ala255ValfsTer24 | p.A255Vfs*24 | Q9BV94 | protein_coding | | | TCGA-BH-A0DP-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Hormone Therapy | arimidex | SD |







 Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells
Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer
Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer
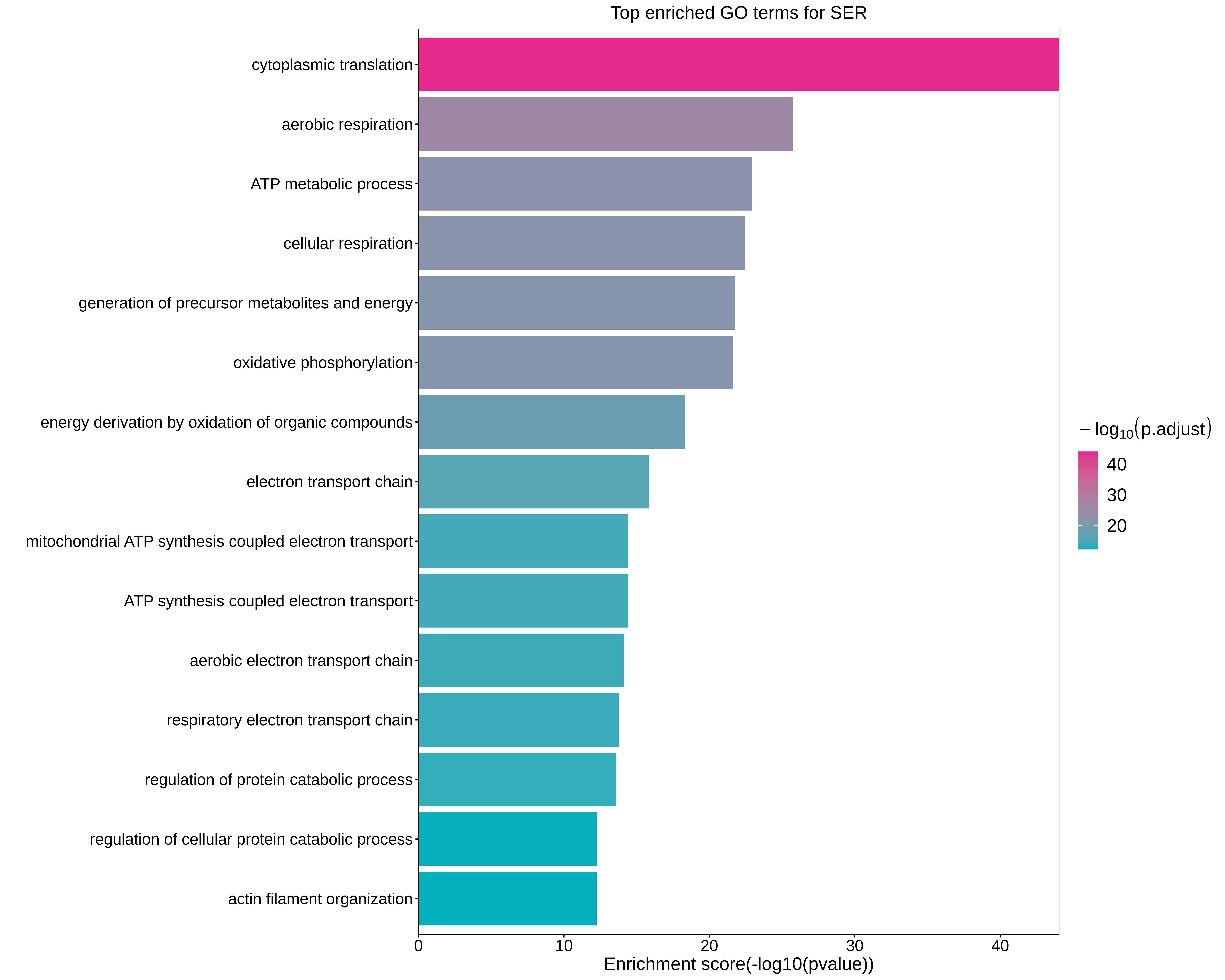

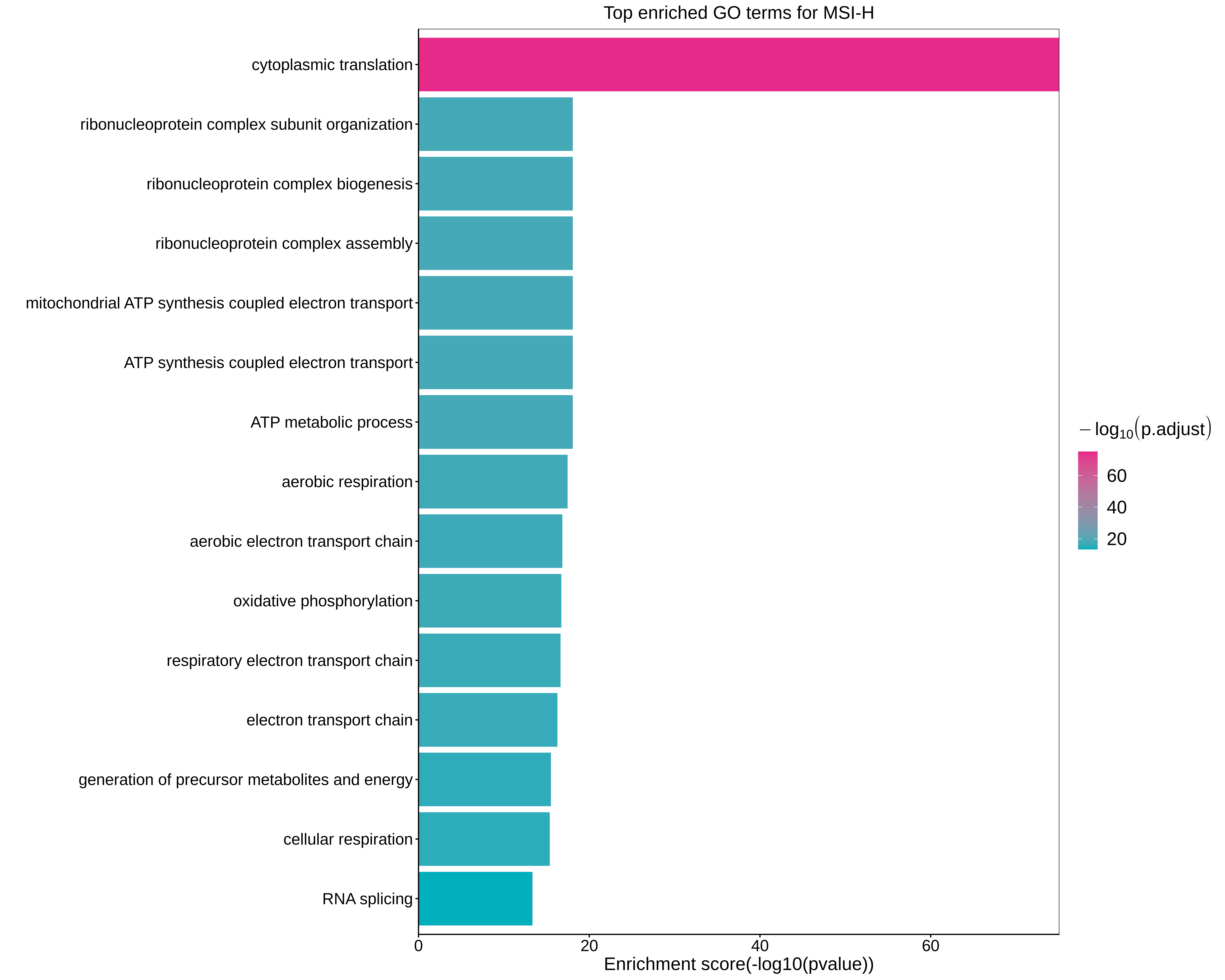
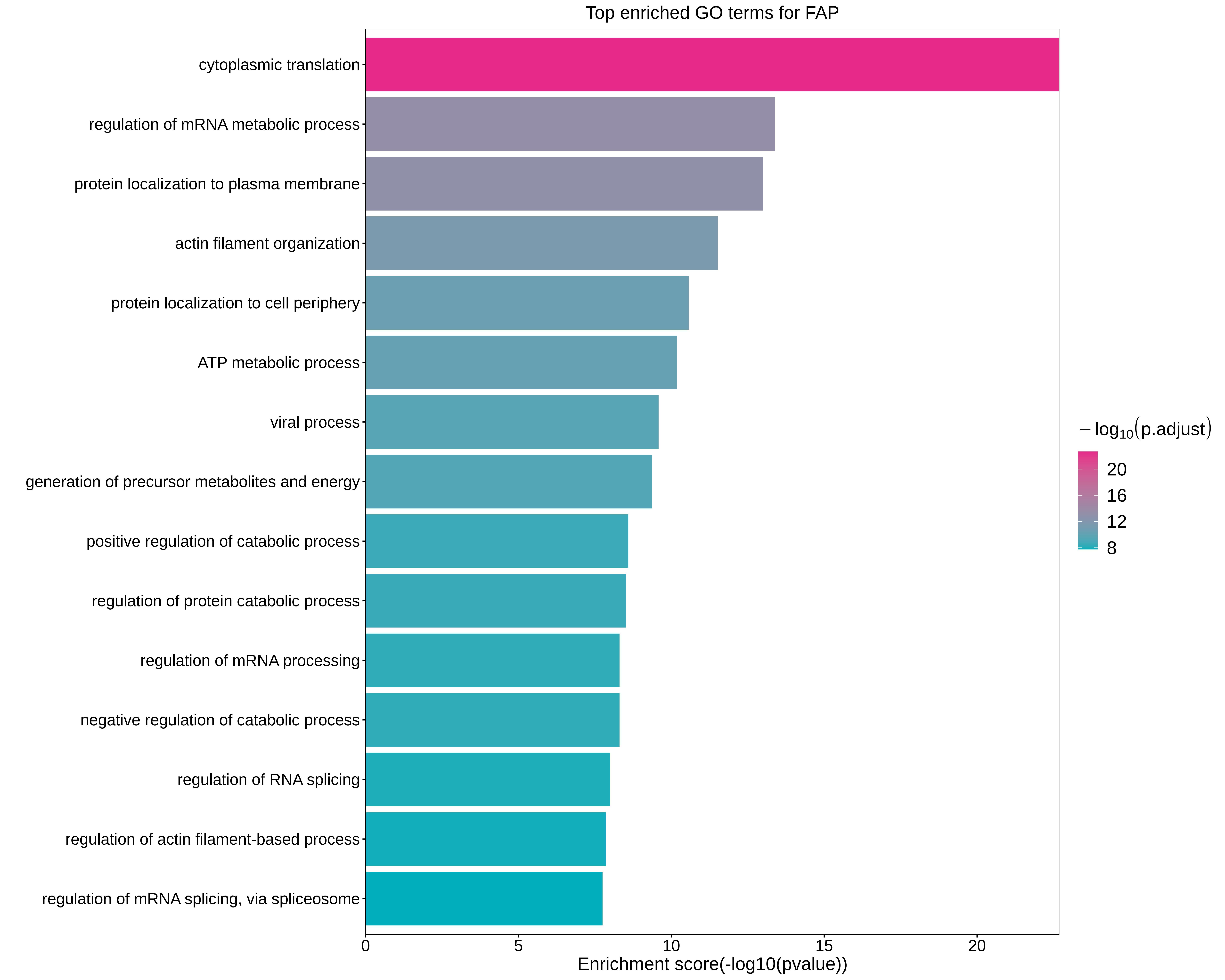
 Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states
Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states
Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states Annotation of somatic variants for genes involved in malignant transformation
Annotation of somatic variants for genes involved in malignant transformation Identification of chemicals and drugs interact with genes involved in malignant transfromation
Identification of chemicals and drugs interact with genes involved in malignant transfromation



