| GO ID | Tissue | Disease Stage | Description | Gene Ratio | Bg Ratio | pvalue | p.adjust | Count |
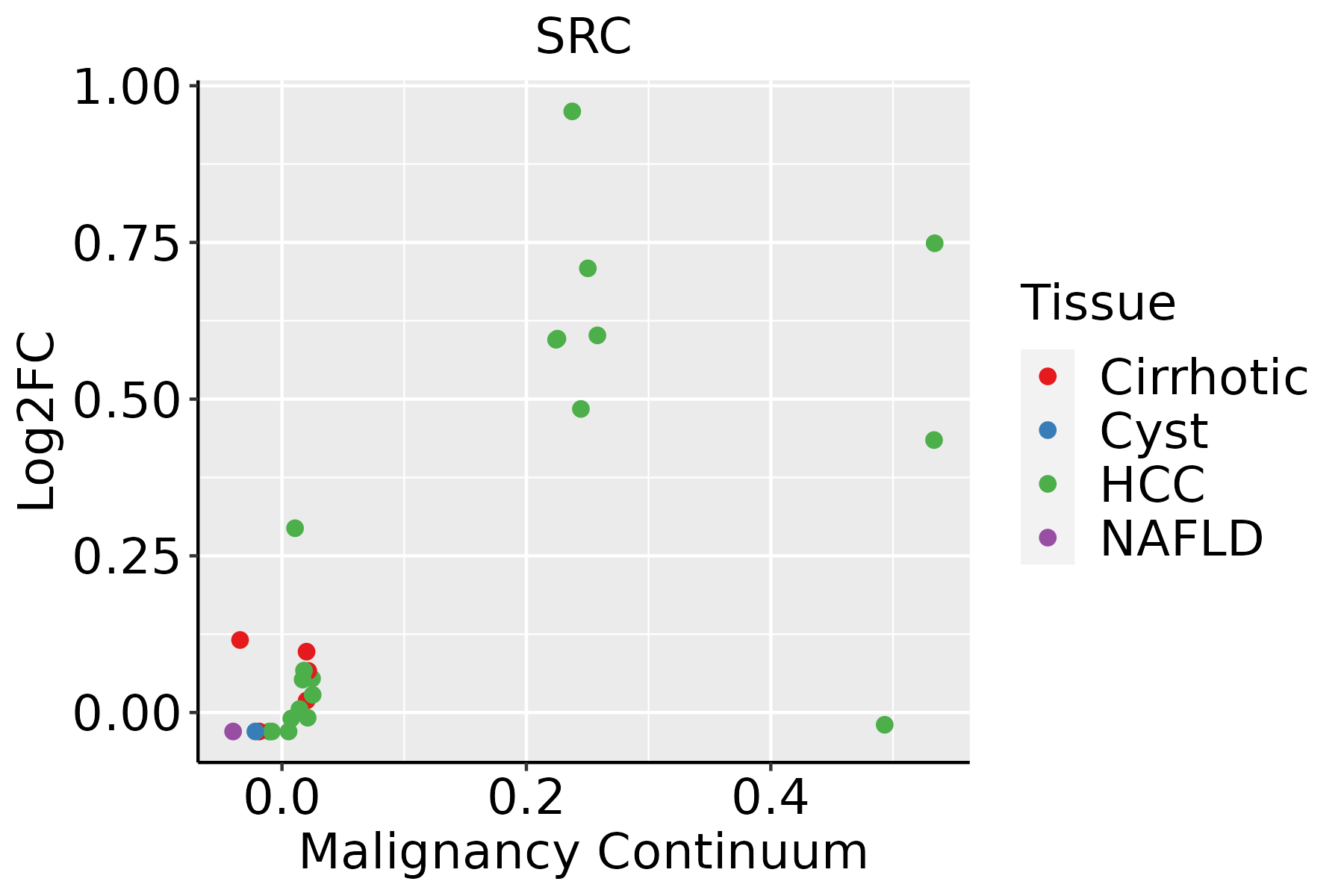
| GO:000155821 | Liver | HCC | regulation of cell growth | 228/7958 | 414/18723 | 1.34e-07 | 2.55e-06 | 228 |
| GO:015011522 | Liver | HCC | cell-substrate junction organization | 69/7958 | 101/18723 | 1.36e-07 | 2.57e-06 | 69 |
| GO:000072311 | Liver | HCC | telomere maintenance | 85/7958 | 131/18723 | 1.86e-07 | 3.40e-06 | 85 |
| GO:200124312 | Liver | HCC | negative regulation of intrinsic apoptotic signaling pathway | 67/7958 | 98/18723 | 1.97e-07 | 3.55e-06 | 67 |
| GO:190285011 | Liver | HCC | microtubule cytoskeleton organization involved in mitosis | 93/7958 | 147/18723 | 2.91e-07 | 5.09e-06 | 93 |
| GO:004328112 | Liver | HCC | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 125/7958 | 209/18723 | 3.12e-07 | 5.42e-06 | 125 |
| GO:000610921 | Liver | HCC | regulation of carbohydrate metabolic process | 109/7958 | 178/18723 | 3.36e-07 | 5.78e-06 | 109 |
| GO:19036493 | Liver | HCC | regulation of cytoplasmic transport | 25/7958 | 28/18723 | 3.42e-07 | 5.88e-06 | 25 |
| GO:00703011 | Liver | HCC | cellular response to hydrogen peroxide | 66/7958 | 98/18723 | 5.74e-07 | 9.07e-06 | 66 |
| GO:000941022 | Liver | HCC | response to xenobiotic stimulus | 248/7958 | 462/18723 | 6.47e-07 | 1.02e-05 | 248 |
| GO:200011612 | Liver | HCC | regulation of cysteine-type endopeptidase activity | 137/7958 | 235/18723 | 6.91e-07 | 1.08e-05 | 137 |
| GO:005105221 | Liver | HCC | regulation of DNA metabolic process | 198/7958 | 359/18723 | 7.62e-07 | 1.17e-05 | 198 |
| GO:005254722 | Liver | HCC | regulation of peptidase activity | 247/7958 | 461/18723 | 8.28e-07 | 1.27e-05 | 247 |
| GO:000704422 | Liver | HCC | cell-substrate junction assembly | 64/7958 | 95/18723 | 8.33e-07 | 1.28e-05 | 64 |
| GO:000166612 | Liver | HCC | response to hypoxia | 172/7958 | 307/18723 | 1.06e-06 | 1.59e-05 | 172 |
| GO:003629312 | Liver | HCC | response to decreased oxygen levels | 179/7958 | 322/18723 | 1.29e-06 | 1.87e-05 | 179 |
| GO:007048212 | Liver | HCC | response to oxygen levels | 191/7958 | 347/18723 | 1.42e-06 | 2.03e-05 | 191 |
| GO:000700412 | Liver | HCC | telomere maintenance via telomerase | 49/7958 | 69/18723 | 1.51e-06 | 2.15e-05 | 49 |
| GO:004315412 | Liver | HCC | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 54/7958 | 78/18723 | 1.61e-06 | 2.27e-05 | 54 |
| GO:005101722 | Liver | HCC | actin filament bundle assembly | 96/7958 | 157/18723 | 1.81e-06 | 2.52e-05 | 96 |
| Pathway ID | Tissue | Disease Stage | Description | Gene Ratio | Bg Ratio | pvalue | p.adjust | qvalue | Count |
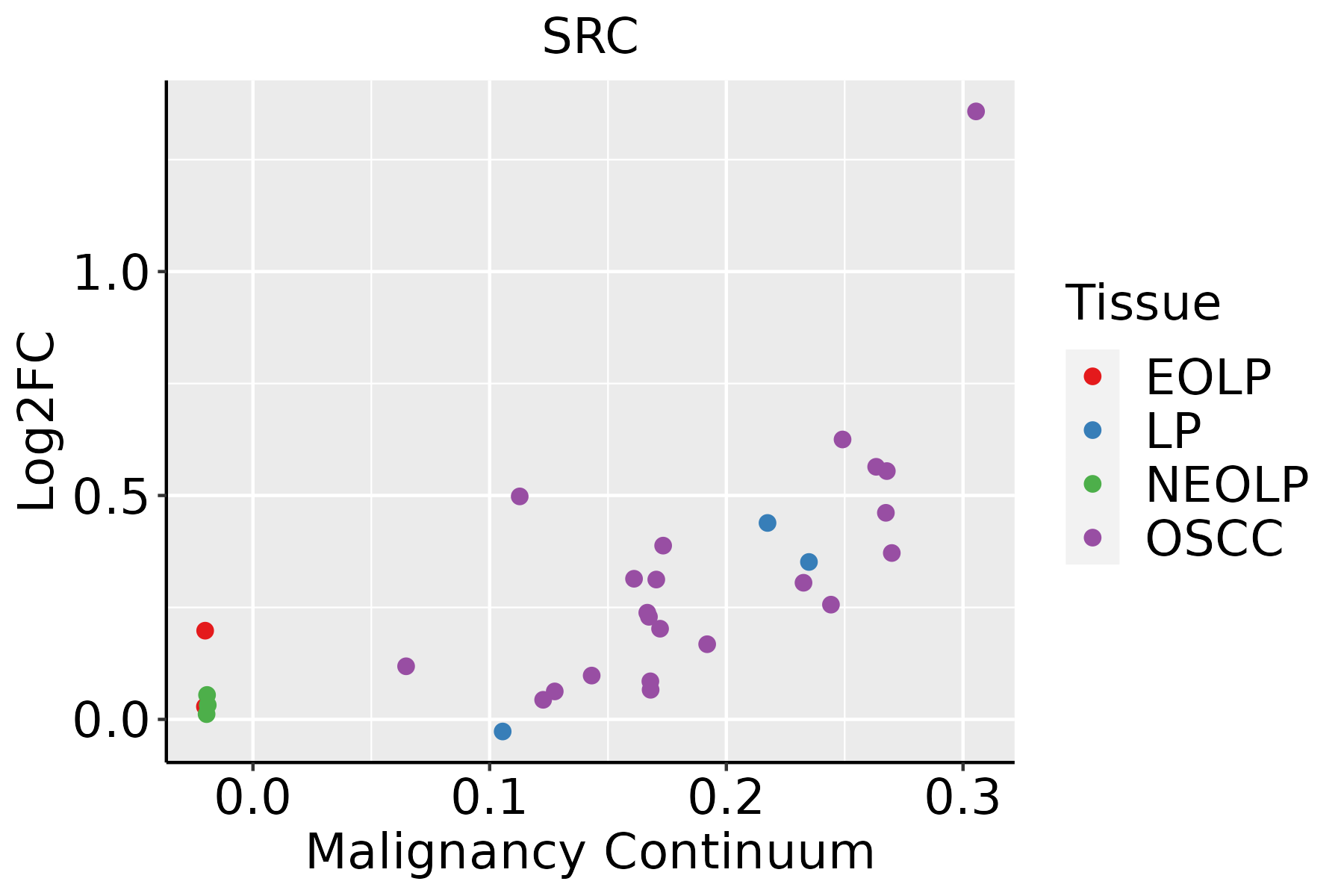
| hsa04144210 | Esophagus | ESCC | Endocytosis | 186/4205 | 251/8465 | 9.74e-16 | 4.66e-14 | 2.39e-14 | 186 |
| hsa05208211 | Esophagus | ESCC | Chemical carcinogenesis - reactive oxygen species | 168/4205 | 223/8465 | 1.83e-15 | 6.81e-14 | 3.49e-14 | 168 |
| hsa05418211 | Esophagus | ESCC | Fluid shear stress and atherosclerosis | 109/4205 | 139/8465 | 2.00e-12 | 3.72e-11 | 1.90e-11 | 109 |
| hsa05131211 | Esophagus | ESCC | Shigellosis | 176/4205 | 247/8465 | 2.27e-12 | 4.01e-11 | 2.05e-11 | 176 |
| hsa05130211 | Esophagus | ESCC | Pathogenic Escherichia coli infection | 142/4205 | 197/8465 | 8.21e-11 | 1.06e-09 | 5.42e-10 | 142 |
| hsa05167211 | Esophagus | ESCC | Kaposi sarcoma-associated herpesvirus infection | 136/4205 | 194/8465 | 4.28e-09 | 4.22e-08 | 2.16e-08 | 136 |
| hsa0513526 | Esophagus | ESCC | Yersinia infection | 100/4205 | 137/8465 | 1.80e-08 | 1.59e-07 | 8.12e-08 | 100 |
| hsa0520529 | Esophagus | ESCC | Proteoglycans in cancer | 138/4205 | 205/8465 | 1.79e-07 | 1.40e-06 | 7.15e-07 | 138 |
| hsa0520325 | Esophagus | ESCC | Viral carcinogenesis | 137/4205 | 204/8465 | 2.47e-07 | 1.88e-06 | 9.62e-07 | 137 |
| hsa05417211 | Esophagus | ESCC | Lipid and atherosclerosis | 143/4205 | 215/8465 | 3.30e-07 | 2.45e-06 | 1.26e-06 | 143 |
| hsa05163210 | Esophagus | ESCC | Human cytomegalovirus infection | 148/4205 | 225/8465 | 5.73e-07 | 4.00e-06 | 2.05e-06 | 148 |
| hsa05100211 | Esophagus | ESCC | Bacterial invasion of epithelial cells | 59/4205 | 77/8465 | 1.05e-06 | 6.76e-06 | 3.46e-06 | 59 |
| hsa0452030 | Esophagus | ESCC | Adherens junction | 69/4205 | 93/8465 | 1.08e-06 | 6.83e-06 | 3.50e-06 | 69 |
| hsa0521910 | Esophagus | ESCC | Bladder cancer | 35/4205 | 41/8465 | 1.91e-06 | 1.15e-05 | 5.87e-06 | 35 |
| hsa051619 | Esophagus | ESCC | Hepatitis B | 108/4205 | 162/8465 | 7.68e-06 | 4.15e-05 | 2.12e-05 | 108 |
| hsa04137210 | Esophagus | ESCC | Mitophagy - animal | 54/4205 | 72/8465 | 9.33e-06 | 4.96e-05 | 2.54e-05 | 54 |
| hsa046259 | Esophagus | ESCC | C-type lectin receptor signaling pathway | 73/4205 | 104/8465 | 1.57e-05 | 7.98e-05 | 4.09e-05 | 73 |
| hsa0512018 | Esophagus | ESCC | Epithelial cell signaling in Helicobacter pylori infection | 52/4205 | 70/8465 | 2.17e-05 | 1.07e-04 | 5.47e-05 | 52 |
| hsa0451020 | Esophagus | ESCC | Focal adhesion | 127/4205 | 203/8465 | 1.25e-04 | 4.99e-04 | 2.56e-04 | 127 |
| hsa0152110 | Esophagus | ESCC | EGFR tyrosine kinase inhibitor resistance | 55/4205 | 79/8465 | 2.44e-04 | 8.78e-04 | 4.50e-04 | 55 |
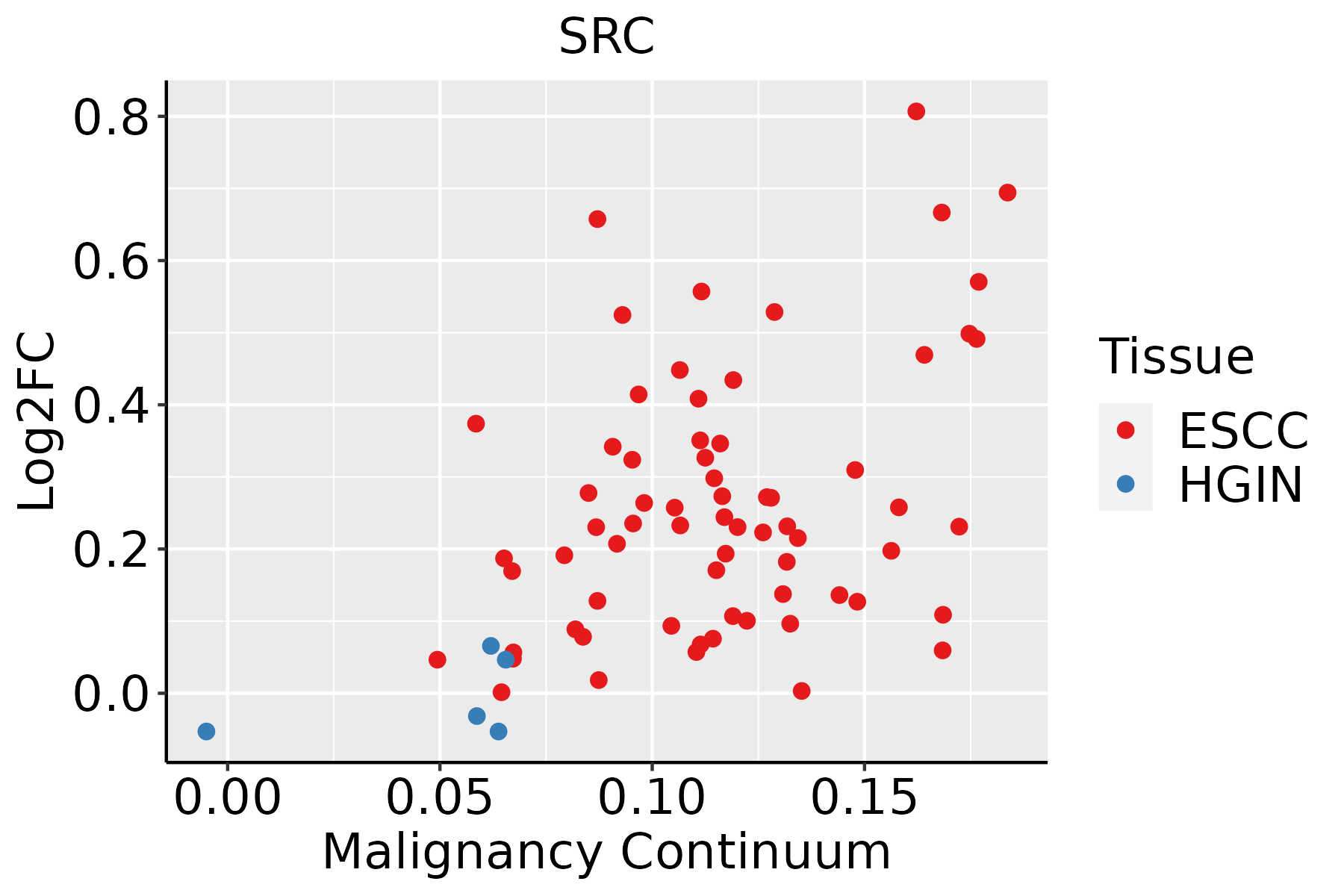
| Hugo Symbol | Variant Class | Variant Classification | dbSNP RS | HGVSc | HGVSp | HGVSp Short | SWISSPROT | BIOTYPE | SIFT | PolyPhen | Tumor Sample Barcode | Tissue | Histology | Sex | Age | Stage | Therapy Types | Drugs | Outcome |
| SRC | SNV | Missense_Mutation | | c.806C>T | p.Ser269Leu | p.S269L | P12931 | protein_coding | deleterious(0) | probably_damaging(0.99) | TCGA-D8-A1J8-01 | Breast | breast invasive carcinoma | Female | >=65 | I/II | Hormone Therapy | nolvadex | SD |
| SRC | insertion | Frame_Shift_Ins | novel | c.1176_1177insCTCATCT | p.Ala393LeufsTer25 | p.A393Lfs*25 | P12931 | protein_coding | | | TCGA-AO-A0JB-01 | Breast | breast invasive carcinoma | Female | <65 | III/IV | Chemotherapy | cyclophosphamide | SD |
| SRC | insertion | Frame_Shift_Ins | novel | c.1177_1178insTGTCTGGGTCCGCTGGGGCCTCTTTC | p.Ala393ValfsTer72 | p.A393Vfs*72 | P12931 | protein_coding | | | TCGA-AO-A0JB-01 | Breast | breast invasive carcinoma | Female | <65 | III/IV | Chemotherapy | cyclophosphamide | SD |
| SRC | SNV | Missense_Mutation | novel | c.11N>G | p.Asn4Ser | p.N4S | P12931 | protein_coding | tolerated_low_confidence(0.97) | benign(0) | TCGA-2W-A8YY-01 | Cervix | cervical & endocervical cancer | Female | <65 | I/II | Chemotherapy | cisplatin | CR |
| SRC | SNV | Missense_Mutation | novel | c.439C>A | p.Gln147Lys | p.Q147K | P12931 | protein_coding | deleterious(0.04) | possibly_damaging(0.882) | TCGA-AA-3947-01 | Colorectum | colon adenocarcinoma | Female | <65 | I/II | Unknown | Unknown | SD |
| SRC | SNV | Missense_Mutation | | c.1571N>T | p.Thr524Met | p.T524M | P12931 | protein_coding | deleterious(0.01) | possibly_damaging(0.52) | TCGA-AD-6899-01 | Colorectum | colon adenocarcinoma | Male | >=65 | III/IV | Unknown | Unknown | SD |
| SRC | SNV | Missense_Mutation | | c.1489N>A | p.Leu497Ile | p.L497I | P12931 | protein_coding | tolerated(1) | probably_damaging(0.993) | TCGA-AD-A5EJ-01 | Colorectum | colon adenocarcinoma | Female | >=65 | I/II | Unknown | Unknown | SD |
| SRC | SNV | Missense_Mutation | novel | c.659N>A | p.Arg220His | p.R220H | P12931 | protein_coding | deleterious(0.01) | benign(0.428) | TCGA-A5-A0G2-01 | Endometrium | uterine corpus endometrioid carcinoma | Female | <65 | III/IV | Unknown | Unknown | SD |
| SRC | SNV | Missense_Mutation | rs533607141 | c.812N>A | p.Arg271Gln | p.R271Q | P12931 | protein_coding | tolerated(0.53) | benign(0.318) | TCGA-AJ-A3BG-01 | Endometrium | uterine corpus endometrioid carcinoma | Female | >=65 | I/II | Chemotherapy | carboplatin | PD |
| SRC | SNV | Missense_Mutation | | c.272C>T | p.Ala91Val | p.A91V | P12931 | protein_coding | deleterious(0.01) | benign(0.437) | TCGA-AP-A059-01 | Endometrium | uterine corpus endometrioid carcinoma | Female | >=65 | I/II | Unknown | Unknown | SD |
| Entrez ID | Symbol | Category | Interaction Types | Drug Claim Name | Drug Name | PMIDs |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | | CEDIRANIB | CEDIRANIB | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | inhibitor | CHEMBL24828 | VANDETANIB | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | inhibitor | CHEMBL3545196 | PD-0166285 | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | | DOVITINIB | DOVITINIB | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | | TYRPHOSTIN A9 | | 11112699 |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | | KX2-361 | KX2-361 | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | | AZD0530 | SARACATINIB | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | inhibitor | 178102599 | | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | inhibitor | CHEMBL1421 | DASATINIB | |
| 6714 | SRC | CLINICALLY ACTIONABLE, KINASE, TYROSINE KINASE, DRUGGABLE GENOME, TRANSCRIPTION FACTOR, ENZYME | inhibitor | 354702286 | | |







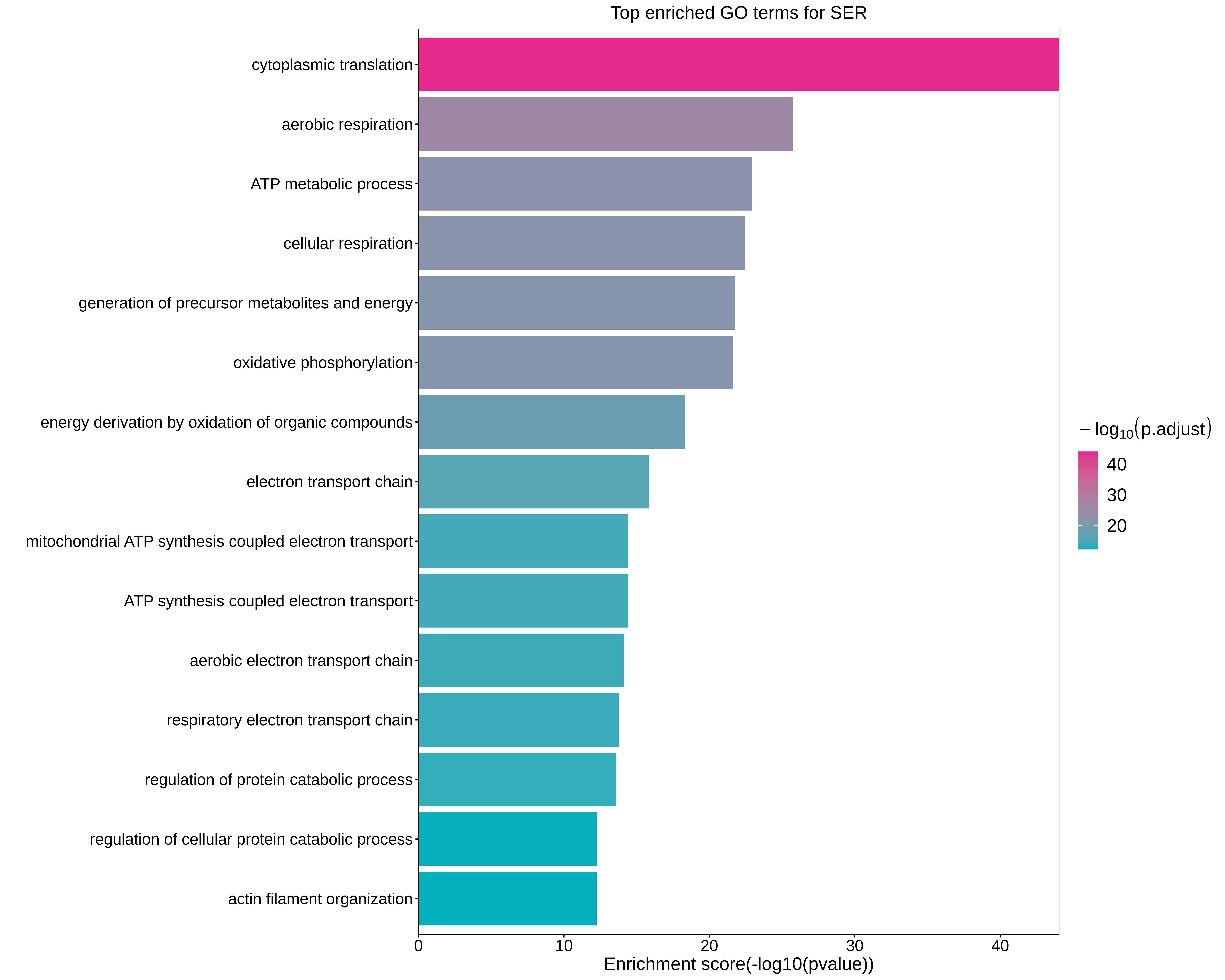
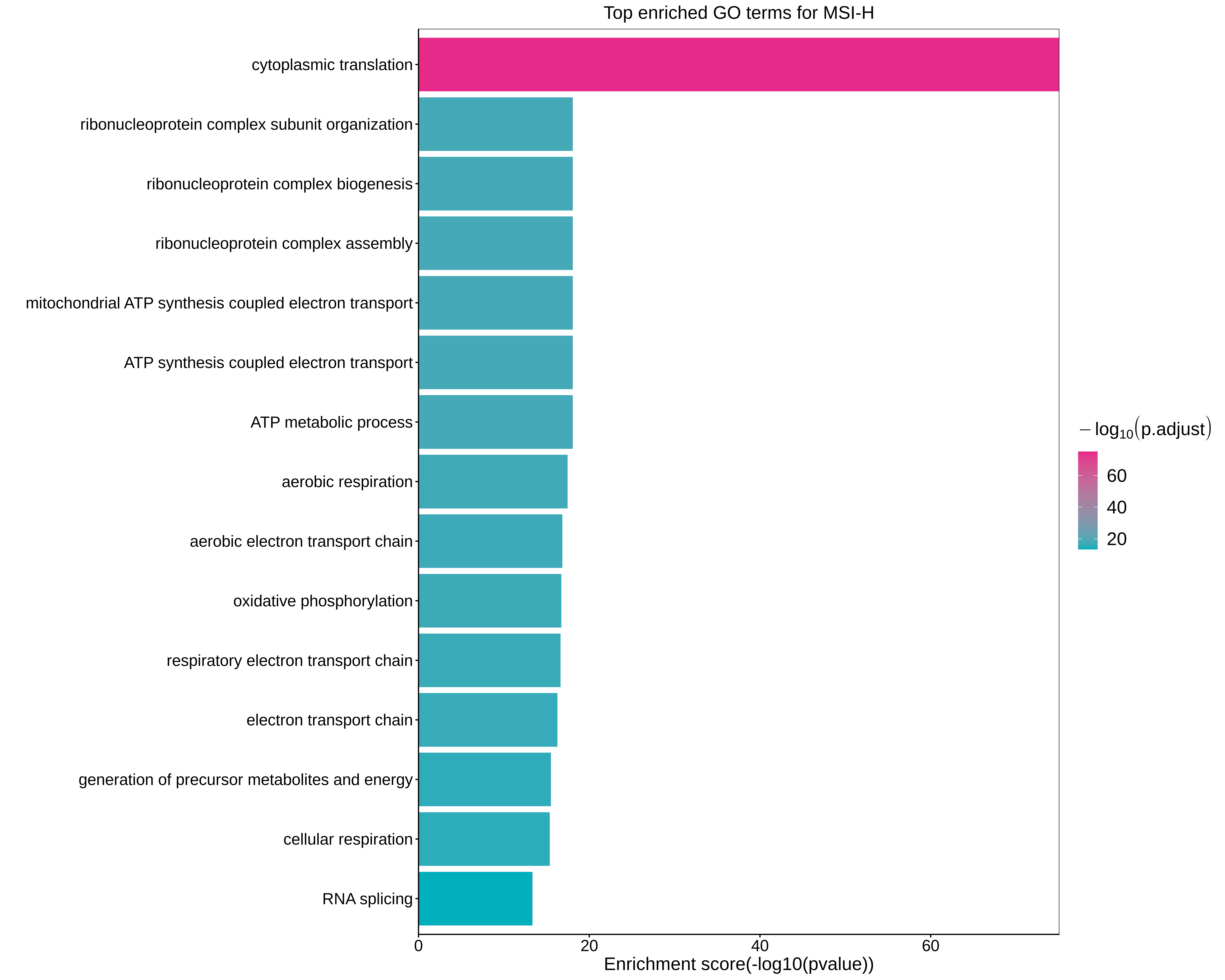
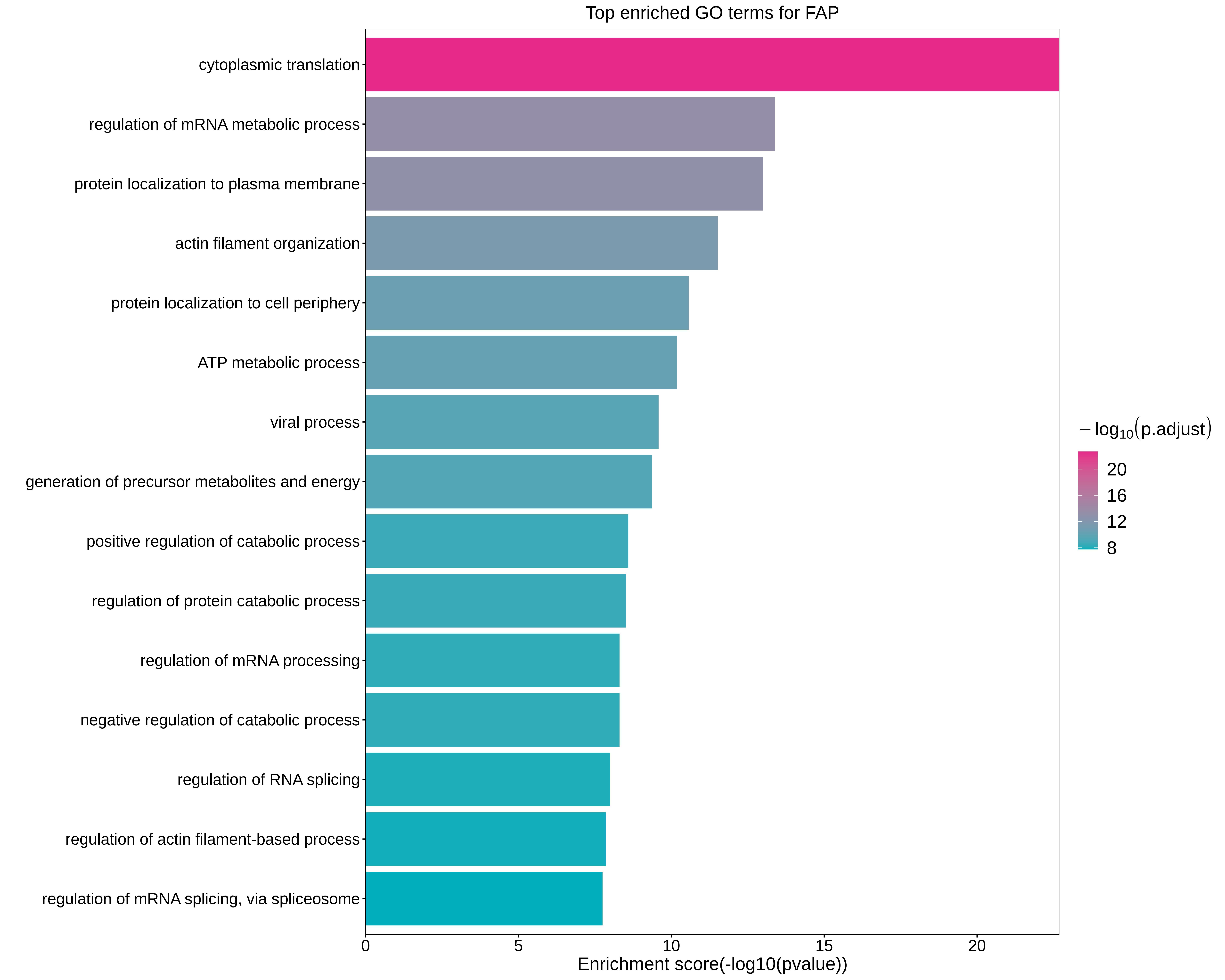
 Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells
Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer
Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer

 Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states
Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states
Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states Annotation of somatic variants for genes involved in malignant transformation
Annotation of somatic variants for genes involved in malignant transformation Identification of chemicals and drugs interact with genes involved in malignant transfromation
Identification of chemicals and drugs interact with genes involved in malignant transfromation