|
|||||
|
 | |
 | |
 | |
 | |
 | |
 | |
 |
Gene: ORAI1 |
Gene summary for ORAI1 |
| Gene information | Species | Human | Gene symbol | ORAI1 | Gene ID | 84876 |
| Gene name | ORAI calcium release-activated calcium modulator 1 | |
| Gene Alias | CRACM1 | |
| Cytomap | 12q24.31 | |
| Gene Type | protein-coding | GO ID | GO:0002115 | UniProtAcc | A0A024RBT3 |
Top |
Malignant transformation analysis |
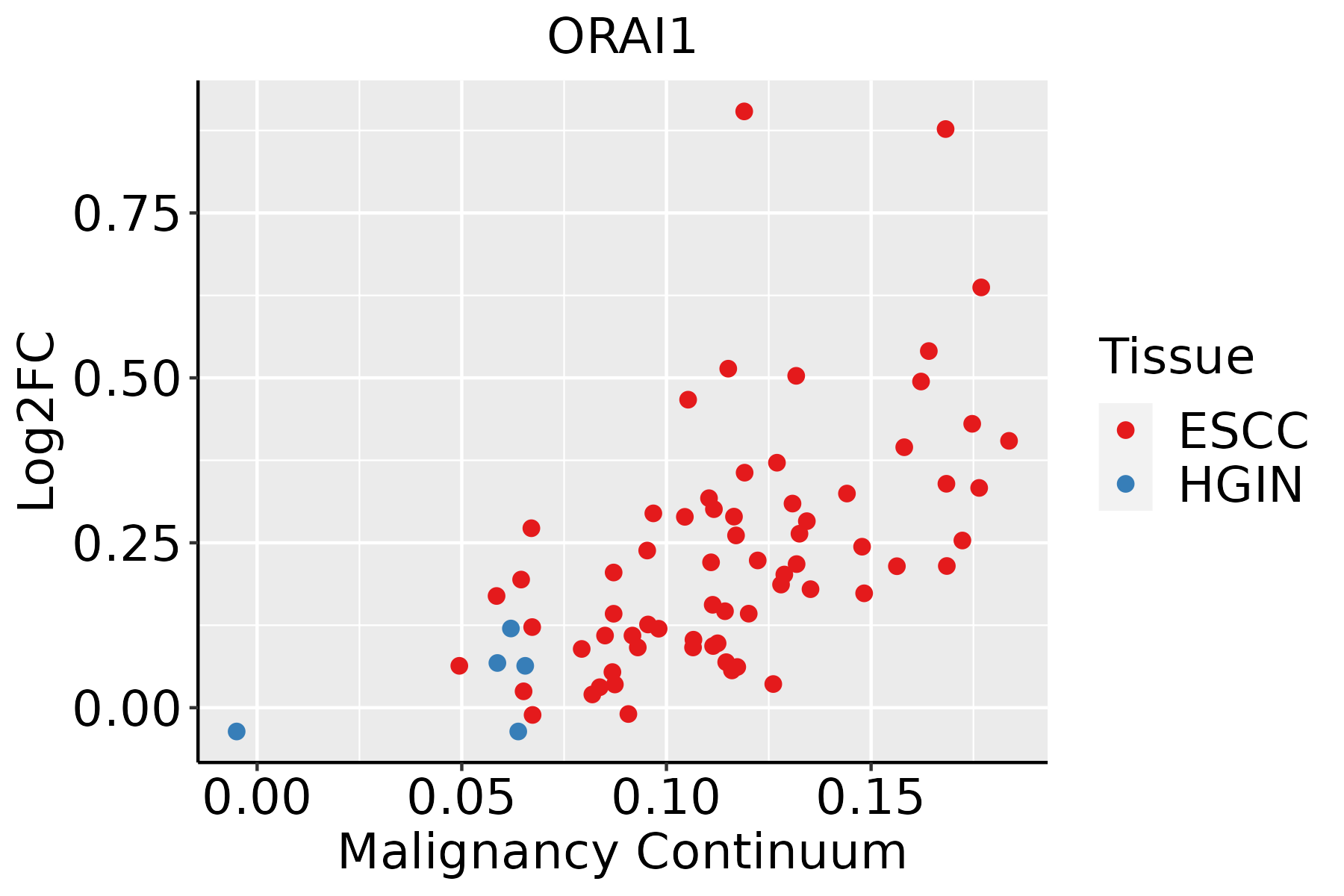
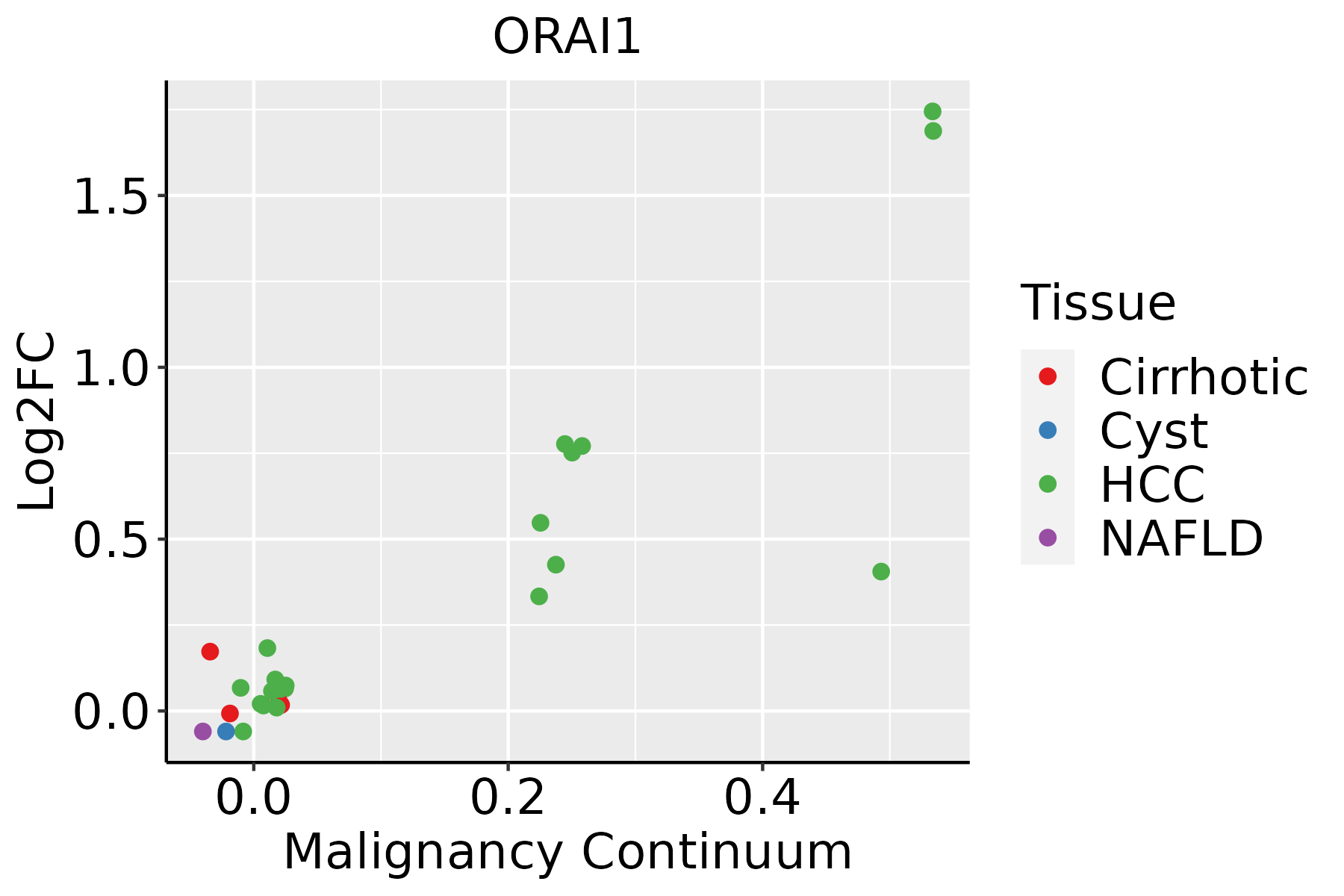
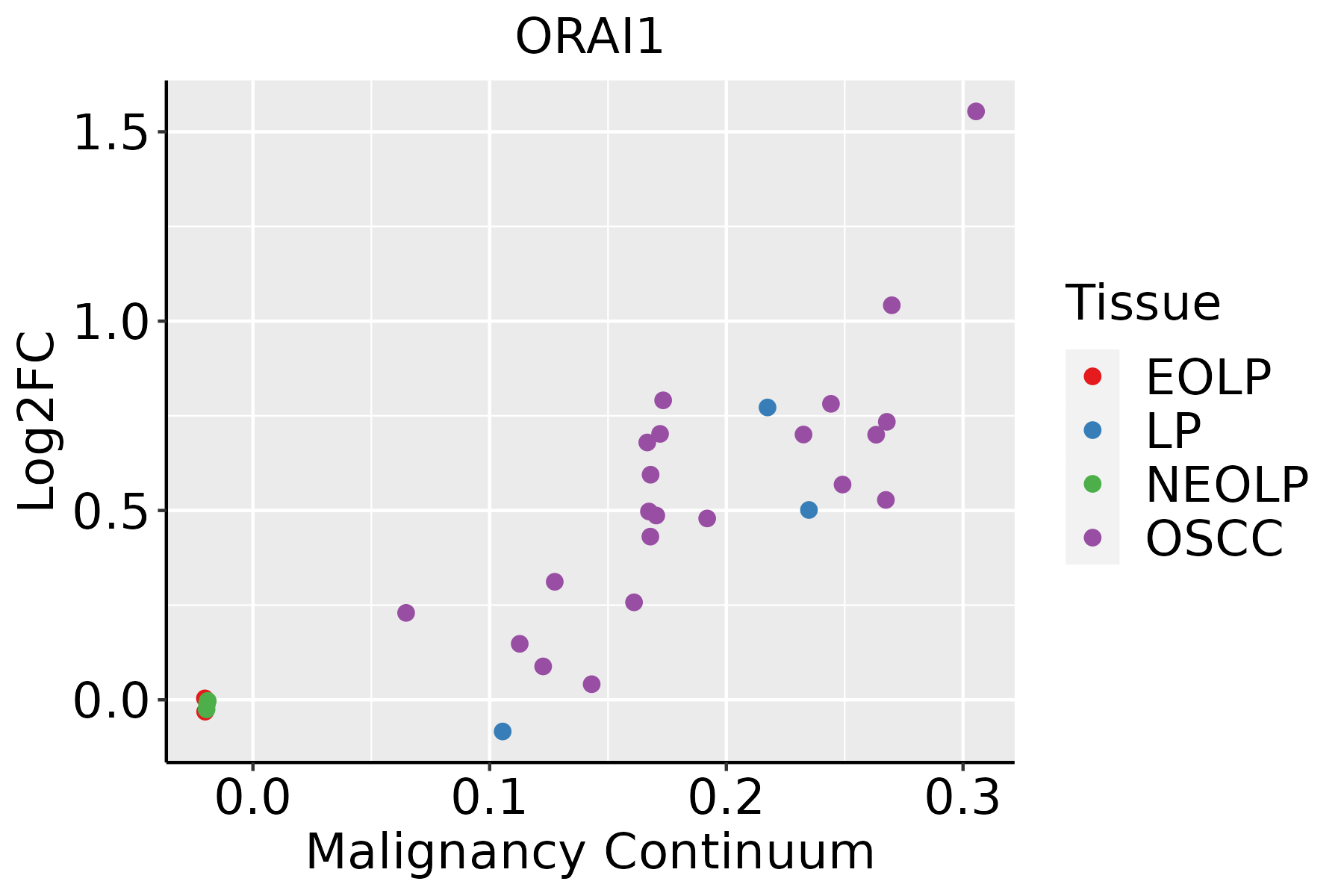
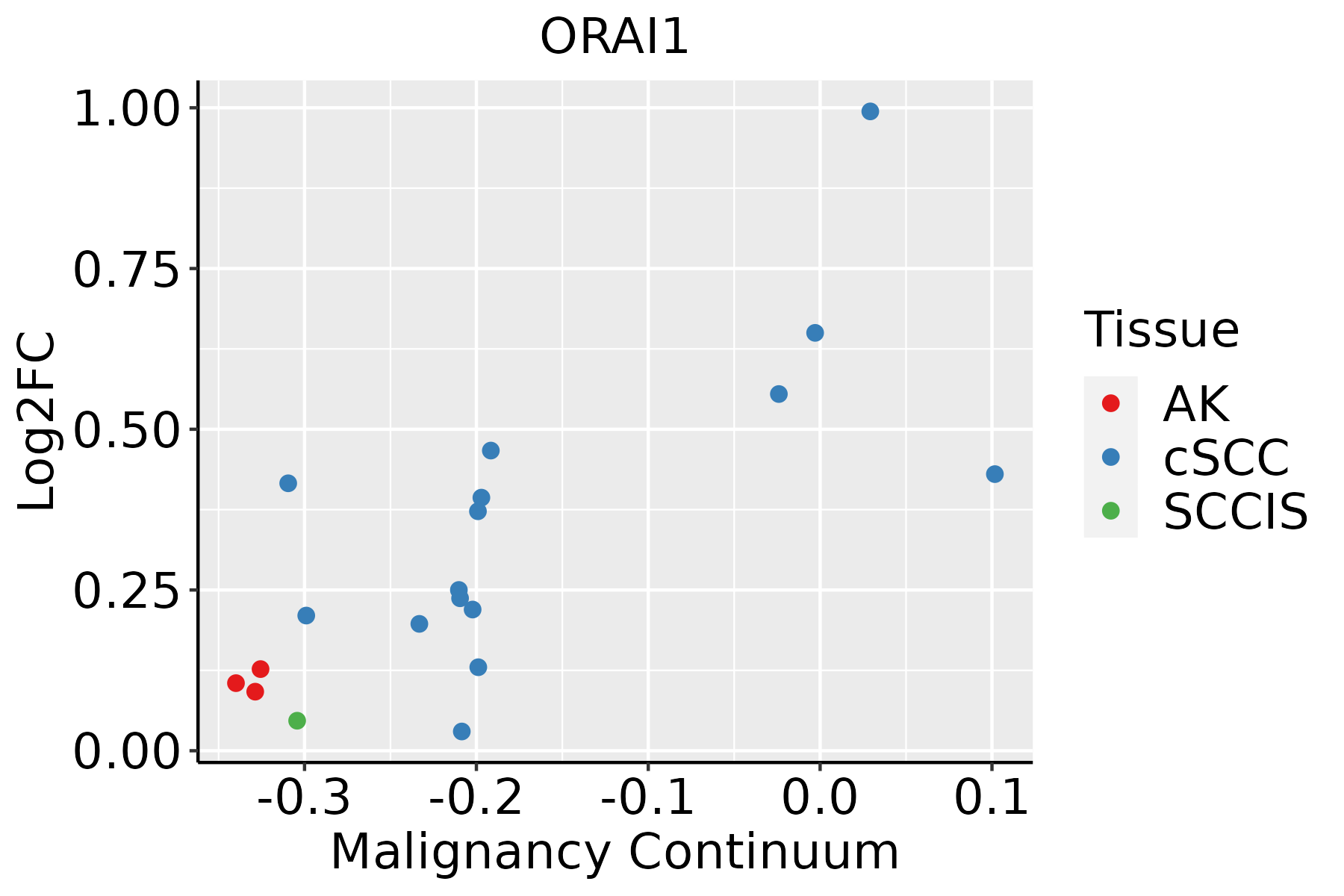
 Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells Identification of the aberrant gene expression in precancerous and cancerous lesions by comparing the gene expression of stem-like cells in diseased tissues with normal stem cells |
| Entrez ID | Symbol | Replicates | Species | Organ | Tissue | Adj P-value | Log2FC | Malignancy |
| 84876 | ORAI1 | P48T-E | Human | Esophagus | ESCC | 2.39e-07 | 1.26e-01 | 0.0959 |
| 84876 | ORAI1 | P49T-E | Human | Esophagus | ESCC | 2.64e-10 | 6.37e-01 | 0.1768 |
| 84876 | ORAI1 | P52T-E | Human | Esophagus | ESCC | 4.28e-22 | 3.95e-01 | 0.1555 |
| 84876 | ORAI1 | P54T-E | Human | Esophagus | ESCC | 1.89e-11 | 2.38e-01 | 0.0975 |
| 84876 | ORAI1 | P56T-E | Human | Esophagus | ESCC | 3.18e-03 | 4.94e-01 | 0.1613 |
| 84876 | ORAI1 | P57T-E | Human | Esophagus | ESCC | 8.48e-05 | 1.09e-01 | 0.0926 |
| 84876 | ORAI1 | P62T-E | Human | Esophagus | ESCC | 6.31e-10 | 2.02e-01 | 0.1302 |
| 84876 | ORAI1 | P65T-E | Human | Esophagus | ESCC | 8.17e-14 | 2.95e-01 | 0.0978 |
| 84876 | ORAI1 | P74T-E | Human | Esophagus | ESCC | 3.62e-04 | 1.73e-01 | 0.1479 |
| 84876 | ORAI1 | P75T-E | Human | Esophagus | ESCC | 1.70e-15 | 2.20e-01 | 0.1125 |
| 84876 | ORAI1 | P76T-E | Human | Esophagus | ESCC | 4.79e-12 | 2.23e-01 | 0.1207 |
| 84876 | ORAI1 | P79T-E | Human | Esophagus | ESCC | 5.80e-05 | 5.65e-02 | 0.1154 |
| 84876 | ORAI1 | P80T-E | Human | Esophagus | ESCC | 5.23e-06 | 2.14e-01 | 0.155 |
| 84876 | ORAI1 | P83T-E | Human | Esophagus | ESCC | 2.14e-12 | 4.30e-01 | 0.1738 |
| 84876 | ORAI1 | P89T-E | Human | Esophagus | ESCC | 2.69e-04 | 3.33e-01 | 0.1752 |
| 84876 | ORAI1 | P91T-E | Human | Esophagus | ESCC | 3.36e-05 | 4.05e-01 | 0.1828 |
| 84876 | ORAI1 | P107T-E | Human | Esophagus | ESCC | 2.99e-12 | 2.53e-01 | 0.171 |
| 84876 | ORAI1 | P127T-E | Human | Esophagus | ESCC | 3.06e-03 | 8.91e-02 | 0.0826 |
| 84876 | ORAI1 | P128T-E | Human | Esophagus | ESCC | 1.29e-11 | 3.56e-01 | 0.1241 |
| 84876 | ORAI1 | P130T-E | Human | Esophagus | ESCC | 4.04e-50 | 8.77e-01 | 0.1676 |
| Page: 1 2 3 4 5 |
| ∗log2FC in expression of this searched gene in stem-like cells from each diseased tissue sample relative to stem-like cells in normal samples in each tissue plotted against the malignancy continuum. Samples are colored based on if they are from different disease stage. |
Top |
Malignant transformation related pathway analysis |
 Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer Find out the enriched GO biological processes and KEGG pathways involved in transition from healthy to precancer to cancer |
| Tissue | Disease Stage | Enriched GO biological Processes |
| Colorectum | AD |  |
| Colorectum | SER | 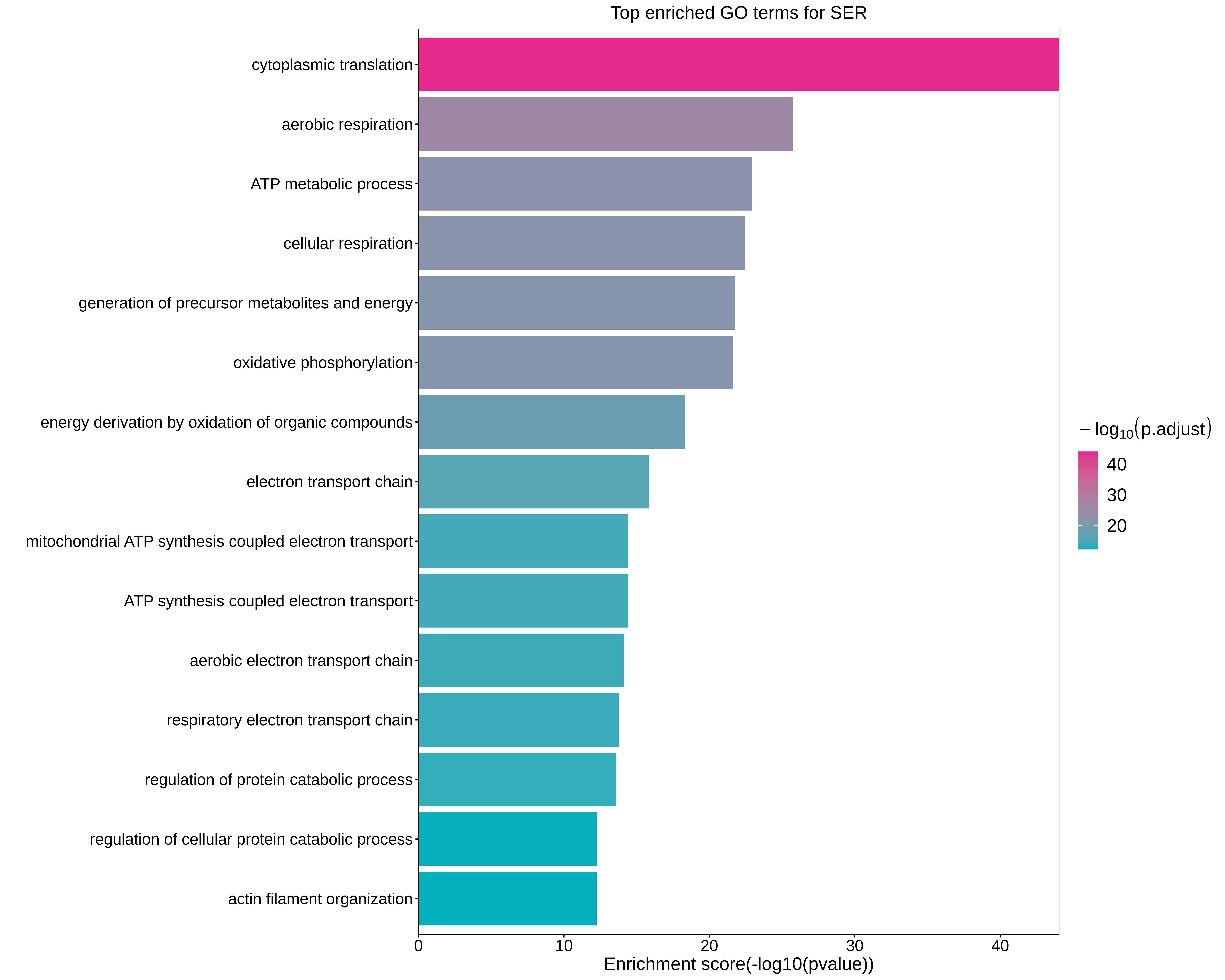 |
| Colorectum | MSS |  |
| Colorectum | MSI-H | 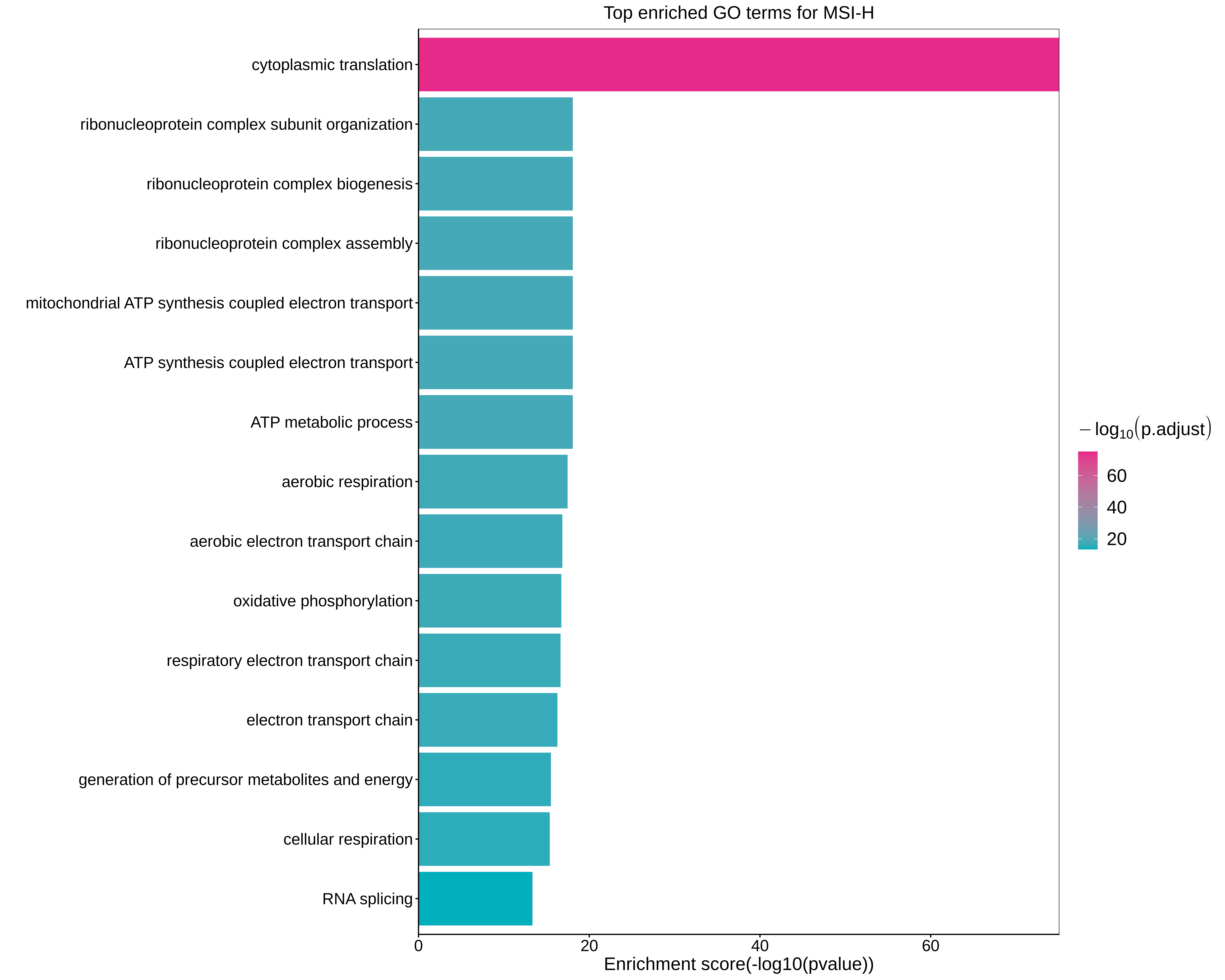 |
| Colorectum | FAP | 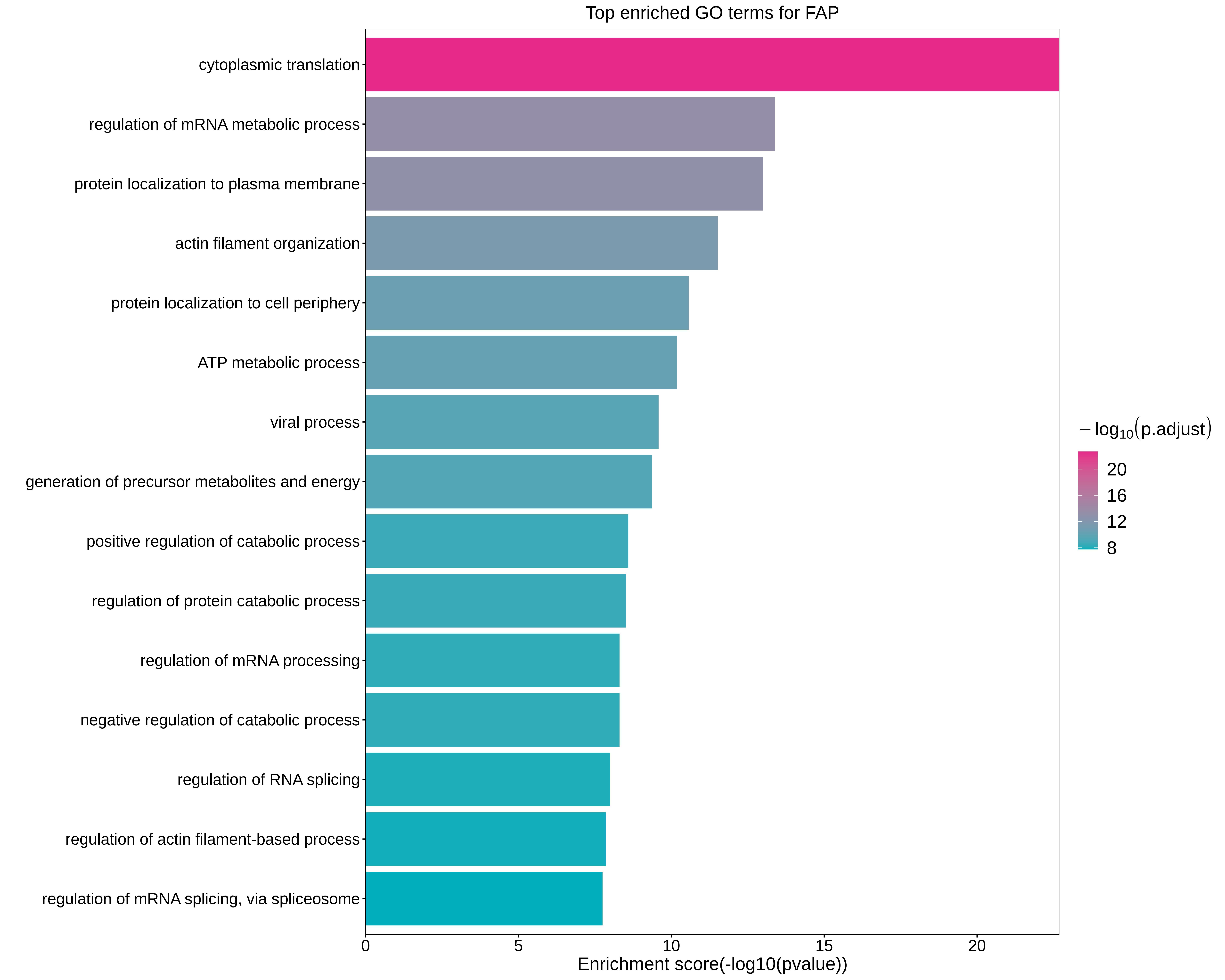 |
| ∗Top 15 enriched GO BP terms are showed in the bar plot of each disease state in each tissue. Each row represents a significant GO biological process which is colored according to the -log10(p.adjust). |
| Page: 1 2 3 4 5 6 7 8 9 |
| GO ID | Tissue | Disease Stage | Description | Gene Ratio | Bg Ratio | pvalue | p.adjust | Count |
| GO:0048732 | Colorectum | AD | gland development | 149/3918 | 436/18723 | 6.07e-11 | 6.33e-09 | 149 |
| GO:0030879 | Colorectum | AD | mammary gland development | 53/3918 | 137/18723 | 1.46e-06 | 4.58e-05 | 53 |
| GO:0061180 | Colorectum | AD | mammary gland epithelium development | 28/3918 | 67/18723 | 8.74e-05 | 1.35e-03 | 28 |
| GO:00487321 | Colorectum | SER | gland development | 112/2897 | 436/18723 | 1.75e-08 | 1.28e-06 | 112 |
| GO:00611801 | Colorectum | SER | mammary gland epithelium development | 22/2897 | 67/18723 | 3.11e-04 | 5.03e-03 | 22 |
| GO:00308791 | Colorectum | SER | mammary gland development | 37/2897 | 137/18723 | 3.61e-04 | 5.61e-03 | 37 |
| GO:00487322 | Colorectum | MSS | gland development | 143/3467 | 436/18723 | 3.85e-13 | 8.01e-11 | 143 |
| GO:00308792 | Colorectum | MSS | mammary gland development | 51/3467 | 137/18723 | 1.88e-07 | 8.26e-06 | 51 |
| GO:00611802 | Colorectum | MSS | mammary gland epithelium development | 28/3467 | 67/18723 | 8.47e-06 | 2.11e-04 | 28 |
| GO:0051349 | Colorectum | MSS | positive regulation of lyase activity | 10/3467 | 23/18723 | 5.06e-03 | 3.47e-02 | 10 |
| GO:00487323 | Colorectum | MSI-H | gland development | 53/1319 | 436/18723 | 7.08e-05 | 2.35e-03 | 53 |
| GO:0048732111 | Esophagus | ESCC | gland development | 269/8552 | 436/18723 | 7.81e-12 | 2.95e-10 | 269 |
| GO:003087914 | Esophagus | ESCC | mammary gland development | 85/8552 | 137/18723 | 8.11e-05 | 6.01e-04 | 85 |
| GO:00611807 | Esophagus | ESCC | mammary gland epithelium development | 43/8552 | 67/18723 | 1.73e-03 | 7.97e-03 | 43 |
| GO:004873222 | Liver | HCC | gland development | 242/7958 | 436/18723 | 2.26e-08 | 5.28e-07 | 242 |
| GO:00308795 | Liver | HCC | mammary gland development | 75/7958 | 137/18723 | 2.52e-03 | 1.22e-02 | 75 |
| GO:004873220 | Oral cavity | OSCC | gland development | 226/7305 | 436/18723 | 2.78e-08 | 5.39e-07 | 226 |
| GO:003087910 | Oral cavity | OSCC | mammary gland development | 69/7305 | 137/18723 | 4.43e-03 | 1.79e-02 | 69 |
| GO:0048732110 | Oral cavity | LP | gland development | 149/4623 | 436/18723 | 4.51e-06 | 9.17e-05 | 149 |
| GO:004873227 | Skin | cSCC | gland development | 167/4864 | 436/18723 | 7.64e-09 | 2.44e-07 | 167 |
| Page: 1 2 |
| Pathway ID | Tissue | Disease Stage | Description | Gene Ratio | Bg Ratio | pvalue | p.adjust | qvalue | Count |
| hsa046112 | Liver | HCC | Platelet activation | 71/4020 | 124/8465 | 1.77e-02 | 4.15e-02 | 2.31e-02 | 71 |
| hsa0461111 | Liver | HCC | Platelet activation | 71/4020 | 124/8465 | 1.77e-02 | 4.15e-02 | 2.31e-02 | 71 |
| Page: 1 |
Top |
Cell-cell communication analysis |
 Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states Identification of potential cell-cell interactions between two cell types and their ligand-receptor pairs for different disease states |
| Ligand | Receptor | LRpair | Pathway | Tissue | Disease Stage |
| Page: 1 |
Top |
Single-cell gene regulatory network inference analysis |
 Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states Find out the significant the regulons (TFs) and the target genes of each regulon across cell types for different disease states |
| TF | Cell Type | Tissue | Disease Stage | Target Gene | RSS | Regulon Activity |
| ∗The dot plots of a searched regulon are shown for all cell subpopulations in each disease state of each tissue based on the regulon specific score inferred using pySCENIC and by calculating the average expression. |
| Page: 1 |
Top |
Somatic mutation of malignant transformation related genes |
 Annotation of somatic variants for genes involved in malignant transformation Annotation of somatic variants for genes involved in malignant transformation |
| Hugo Symbol | Variant Class | Variant Classification | dbSNP RS | HGVSc | HGVSp | HGVSp Short | SWISSPROT | BIOTYPE | SIFT | PolyPhen | Tumor Sample Barcode | Tissue | Histology | Sex | Age | Stage | Therapy Types | Drugs | Outcome |
| ORAI1 | SNV | Missense_Mutation | novel | c.709N>C | p.Tyr237His | p.Y237H | protein_coding | deleterious_low_confidence(0) | benign(0.185) | TCGA-D8-A1XK-01 | Breast | breast invasive carcinoma | Female | <65 | I/II | Chemotherapy | doxorubicine+cyclophosphamide | SD | |
| ORAI1 | deletion | Frame_Shift_Del | novel | c.672delN | p.His225ThrfsTer7 | p.H225Tfs*7 | protein_coding | TCGA-EW-A2FV-01 | Breast | breast invasive carcinoma | Female | <65 | III/IV | Chemotherapy | docetaxel | SD | |||
| ORAI1 | SNV | Missense_Mutation | rs782280709 | c.251A>G | p.Asn84Ser | p.N84S | protein_coding | deleterious(0.03) | probably_damaging(0.926) | TCGA-ZJ-AAX4-01 | Cervix | cervical & endocervical cancer | Female | >=65 | I/II | Unknown | Unknown | SD | |
| ORAI1 | SNV | Missense_Mutation | novel | c.614G>A | p.Arg205Gln | p.R205Q | protein_coding | tolerated(0.08) | probably_damaging(0.978) | TCGA-A6-2679-01 | Colorectum | colon adenocarcinoma | Female | >=65 | I/II | Unknown | Unknown | SD | |
| ORAI1 | SNV | Missense_Mutation | novel | c.427N>T | p.Pro143Ser | p.P143S | protein_coding | tolerated_low_confidence(0.77) | benign(0.003) | TCGA-A6-2686-01 | Colorectum | colon adenocarcinoma | Female | >=65 | I/II | Unknown | Unknown | SD | |
| ORAI1 | SNV | Missense_Mutation | rs782538669 | c.310N>T | p.Arg104Cys | p.R104C | protein_coding | deleterious(0) | probably_damaging(0.999) | TCGA-A6-A565-01 | Colorectum | colon adenocarcinoma | Female | <65 | III/IV | Unspecific | 5FU | PD | |
| ORAI1 | SNV | Missense_Mutation | rs368310115 | c.221N>T | p.Ala74Val | p.A74V | protein_coding | deleterious(0) | possibly_damaging(0.869) | TCGA-AA-3821-01 | Colorectum | colon adenocarcinoma | Female | >=65 | I/II | Unknown | Unknown | SD | |
| ORAI1 | SNV | Missense_Mutation | novel | c.481N>A | p.Val161Ile | p.V161I | protein_coding | tolerated(0.27) | benign(0.017) | TCGA-AA-3864-01 | Colorectum | colon adenocarcinoma | Male | >=65 | I/II | Unknown | Unknown | SD | |
| ORAI1 | SNV | Missense_Mutation | novel | c.636N>T | p.Glu212Asp | p.E212D | protein_coding | tolerated(0.07) | probably_damaging(0.978) | TCGA-G4-6588-01 | Colorectum | colon adenocarcinoma | Female | <65 | I/II | Unknown | Unknown | SD | |
| ORAI1 | SNV | Missense_Mutation | novel | c.686A>C | p.His229Pro | p.H229P | protein_coding | tolerated(0.25) | benign(0) | TCGA-A5-A0GP-01 | Endometrium | uterine corpus endometrioid carcinoma | Female | <65 | I/II | Unknown | Unknown | SD |
| Page: 1 2 3 |
Top |
Related drugs of malignant transformation related genes |
 Identification of chemicals and drugs interact with genes involved in malignant transfromation Identification of chemicals and drugs interact with genes involved in malignant transfromation |
| (DGIdb 4.0) |
| Entrez ID | Symbol | Category | Interaction Types | Drug Claim Name | Drug Name | PMIDs |
| 84876 | ORAI1 | TRANSCRIPTION FACTOR, ION CHANNEL, TRANSPORTER | CM2489 | |||
| 84876 | ORAI1 | TRANSCRIPTION FACTOR, ION CHANNEL, TRANSPORTER | blocker | 381118797 | ||
| 84876 | ORAI1 | TRANSCRIPTION FACTOR, ION CHANNEL, TRANSPORTER | blocker | 381118798 | ||
| 84876 | ORAI1 | TRANSCRIPTION FACTOR, ION CHANNEL, TRANSPORTER | RP4010 | |||
| 84876 | ORAI1 | TRANSCRIPTION FACTOR, ION CHANNEL, TRANSPORTER | blocker | 375973230 | ||
| 84876 | ORAI1 | TRANSCRIPTION FACTOR, ION CHANNEL, TRANSPORTER | blocker | 375973229 |
| Page: 1 |