|
||||||||
|
||||||||
Gene summary for ICAM2 |
| Gene Symbol | ICAM2 | Gene ID | 3384 |
| Gene name | intercellular adhesion molecule 2 |
| Synonyms | CD102 |
| Type of gene | protein_coding |
| UniProtAcc | P13598 |
| GO ID | GO term |
| GO:0007155 | cell adhesion |
| GO:0098609 | cell-cell adhesion |
Top |
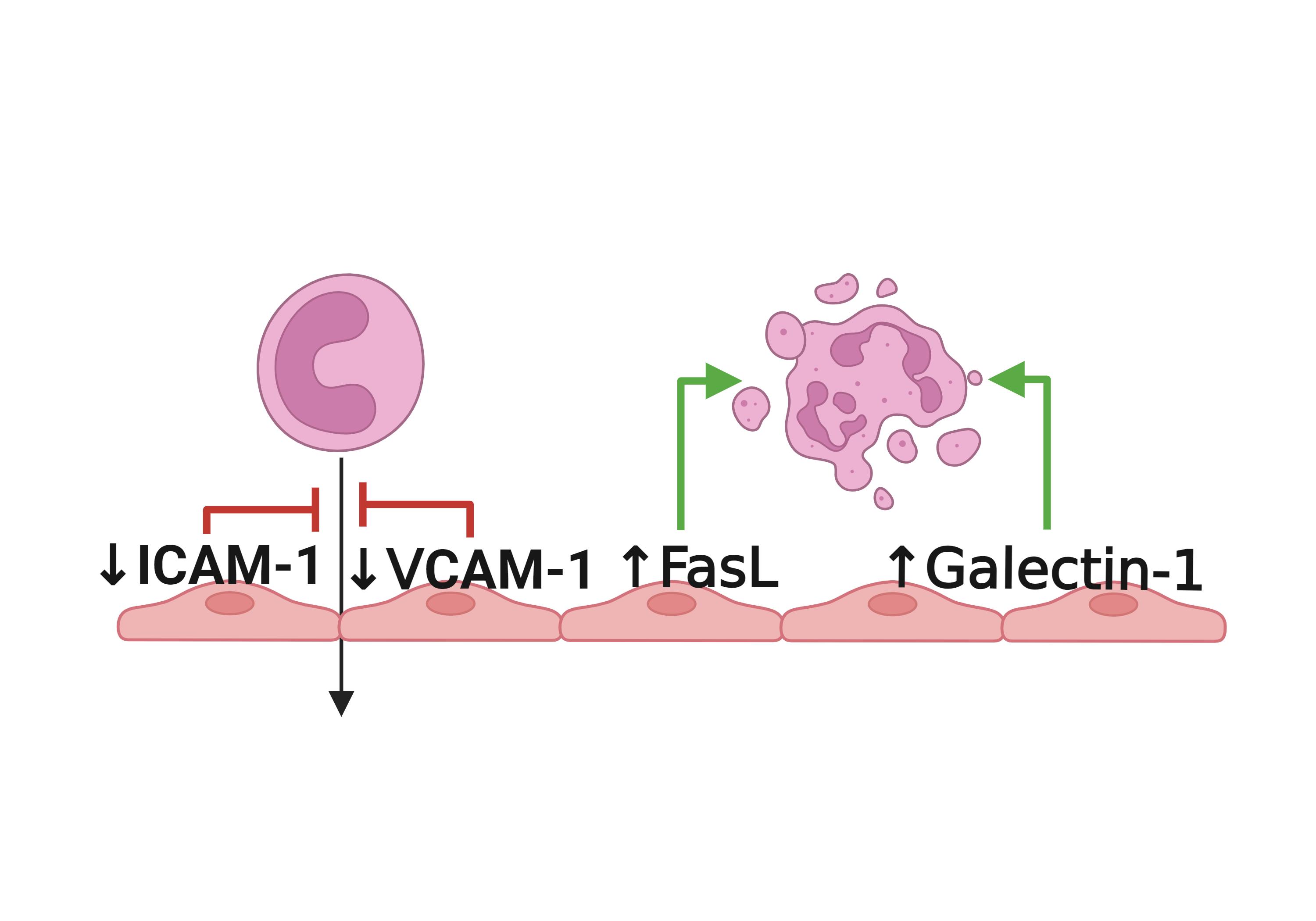
Literatures describing the association of ICAM2 and immune escape mechanisms |
Top |
Comparison of the ICAM2 expression level between tumor and normal groups |
Top |
Comparison of the ICAM2 methylation level between tumor and normal groups |
Top |
Summary of the copy number in TCGA tumor samples |
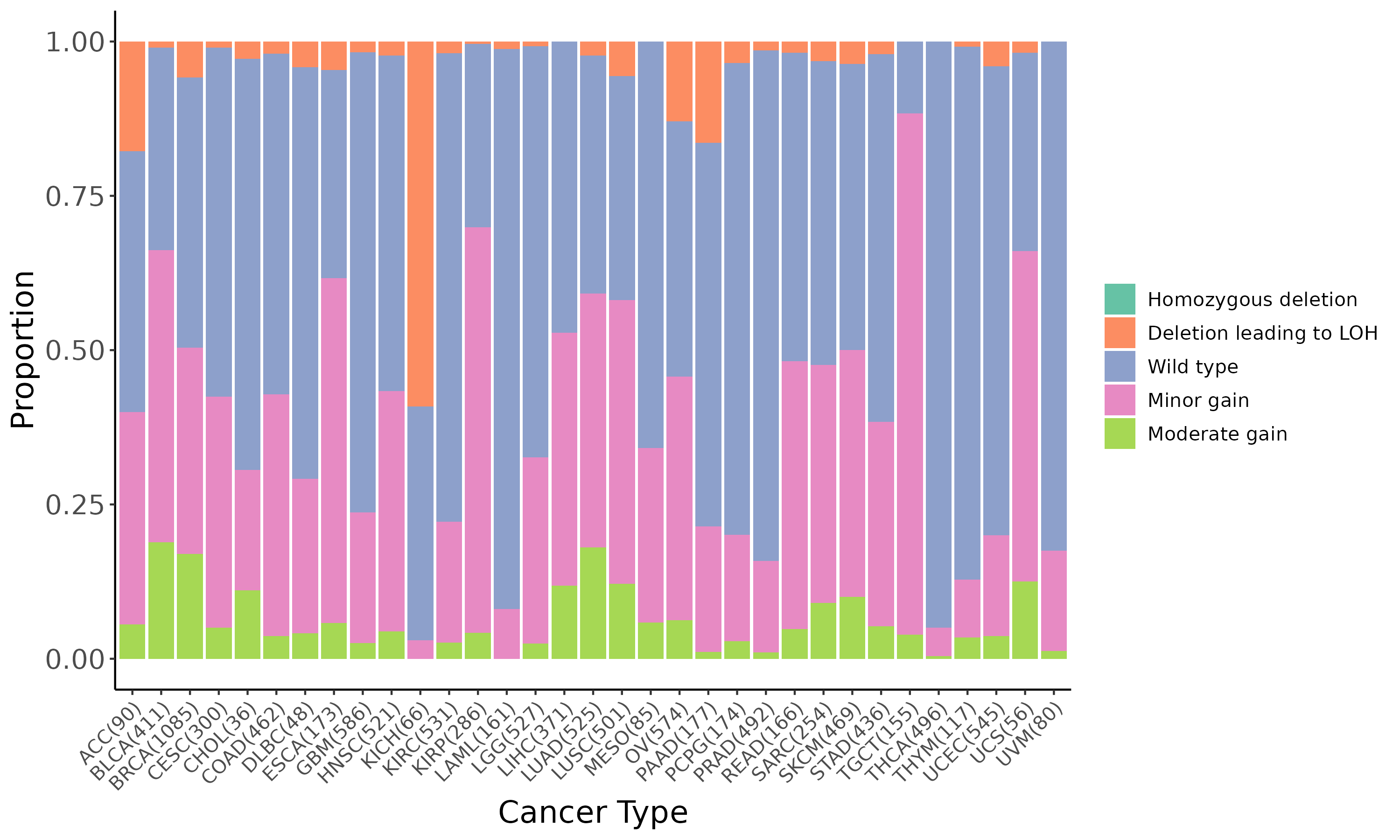 |
Top |
DEGs and the enrichment analysis between the mutated and wild type groups |
| Gene ID | Symbol | Log2 Fold Change | P-value | Adjusted P-value |
|---|---|---|---|---|
| ENSG00000205436 | EXOC3L4 | -2.41e+00 | 1.21e-04 | 9.55e-03 |
| ENSG00000253771 | TPTE2P1 | -1.84e+00 | 1.22e-04 | 9.61e-03 |
| ENSG00000064763 | FAR2 | -1.57e+00 | 1.23e-04 | 9.61e-03 |
| ENSG00000095932 | SMIM24 | -2.47e+00 | 1.23e-04 | 9.61e-03 |
| ENSG00000132554 | RGS22 | -2.78e+00 | 1.24e-04 | 9.61e-03 |
| ENSG00000189045 | ANKDD1B | -2.14e+00 | 1.23e-04 | 9.61e-03 |
| ENSG00000244564 | RP11-14D22.1 | -3.53e+00 | 1.24e-04 | 9.61e-03 |
| ENSG00000283646 | RP11-24F11.5 | -2.41e+00 | 1.24e-04 | 9.61e-03 |
| ENSG00000160862 | AZGP1 | -3.00e+00 | 1.25e-04 | 9.68e-03 |
| ENSG00000231412 | CTC-490G23.2 | -3.48e+00 | 1.26e-04 | 9.68e-03 |
| ENSG00000203523 | TAS2R2P | -3.08e+00 | 1.27e-04 | 9.73e-03 |
| ENSG00000021300 | PLEKHB1 | -1.46e+00 | 1.28e-04 | 9.81e-03 |
| ENSG00000134757 | DSG3 | -4.10e+00 | 1.30e-04 | 9.92e-03 |
| ENSG00000111834 | RSPH4A | -2.41e+00 | 1.32e-04 | 9.93e-03 |
| ENSG00000170703 | TTLL6 | -2.47e+00 | 1.32e-04 | 9.93e-03 |
| ENSG00000177468 | OLIG3 | -4.50e+00 | 1.32e-04 | 9.93e-03 |
| ENSG00000184985 | SORCS2 | -2.22e+00 | 1.32e-04 | 9.93e-03 |
| ENSG00000213981 | AC007277.3 | -3.01e+00 | 1.32e-04 | 9.93e-03 |
| ENSG00000264449 | RP11-945C19.4 | -3.47e+00 | 1.32e-04 | 9.93e-03 |
| ENSG00000102891 | MT4 | -7.42e+00 | 1.33e-04 | 9.96e-03 |
| Page: 1 2 ... 16 17 18 19 20 ... 50 51 |
Down-regulated KEGG pathways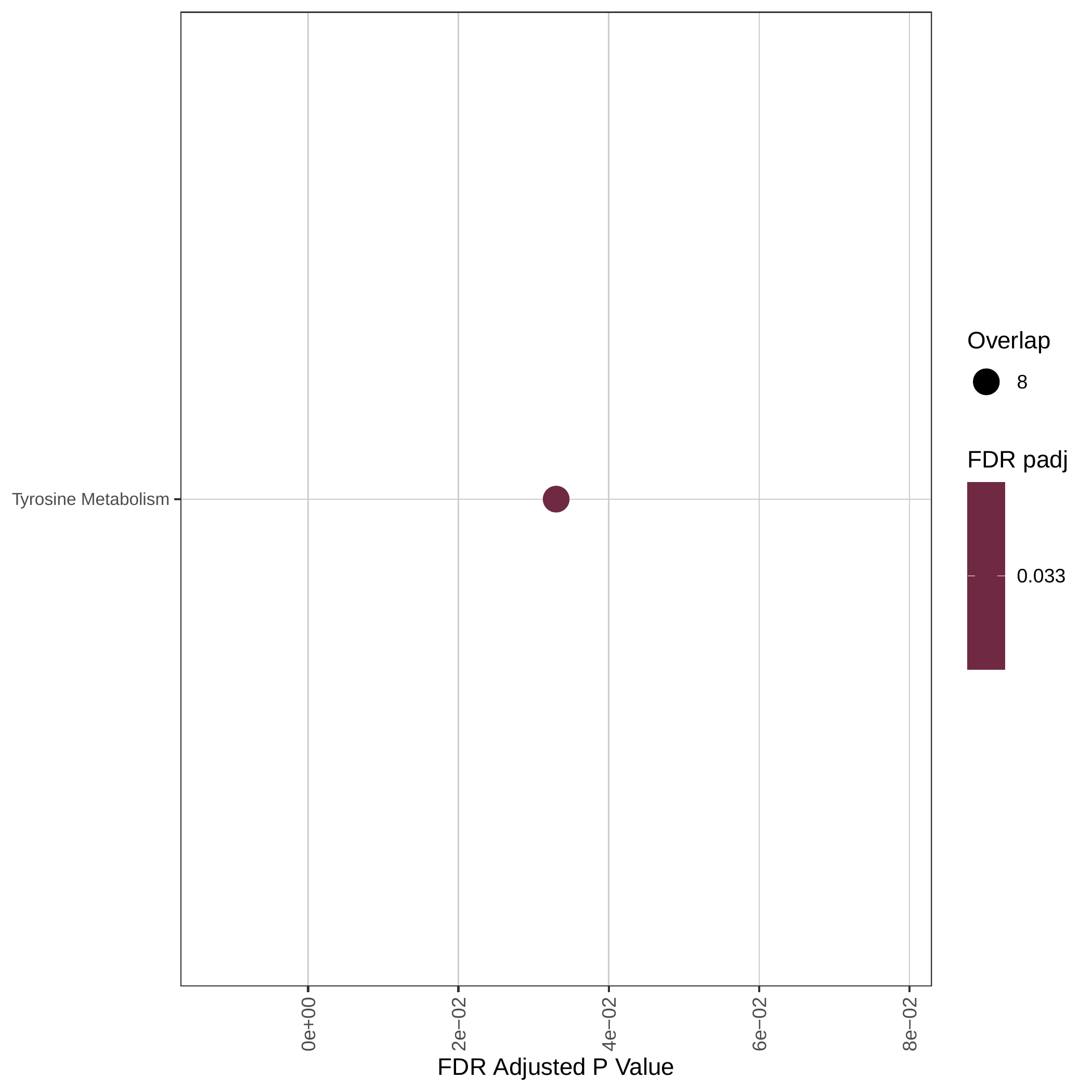 | Down-regulated GOBP pathways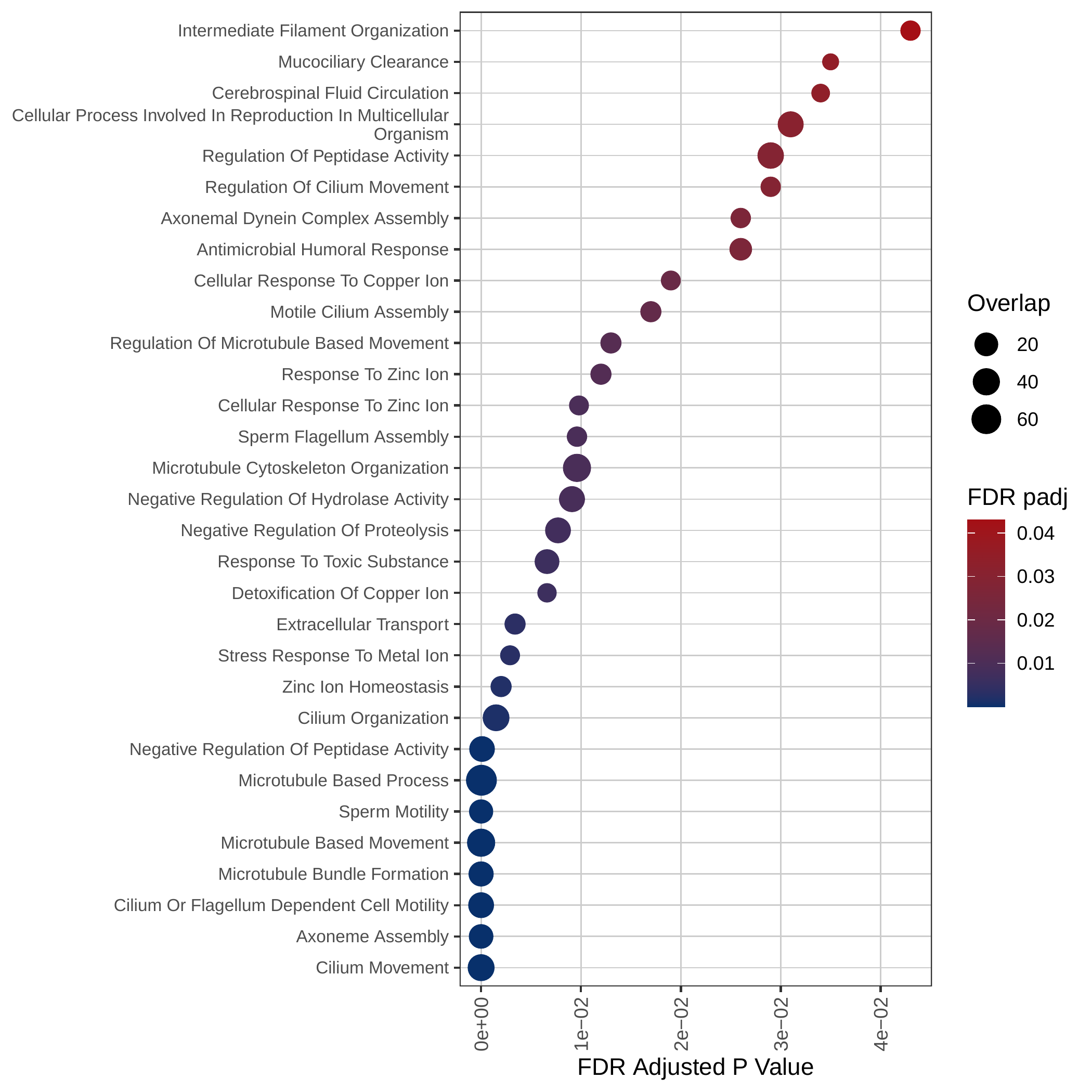 | Down-regulated Hallmark pathways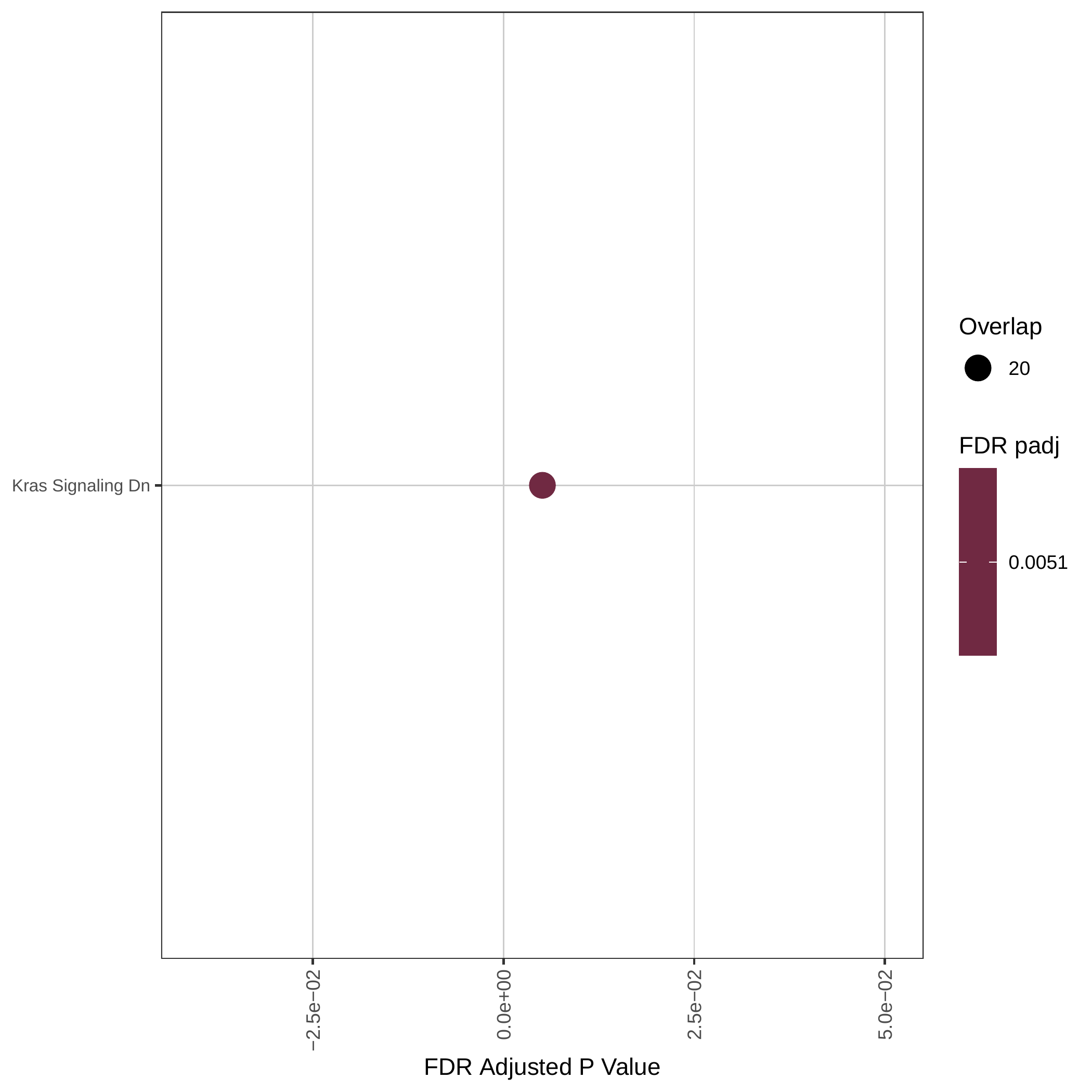 |
Gene expression and mutation differences between non-responders and responders after immunotherapy |
| Expression | Mutation |
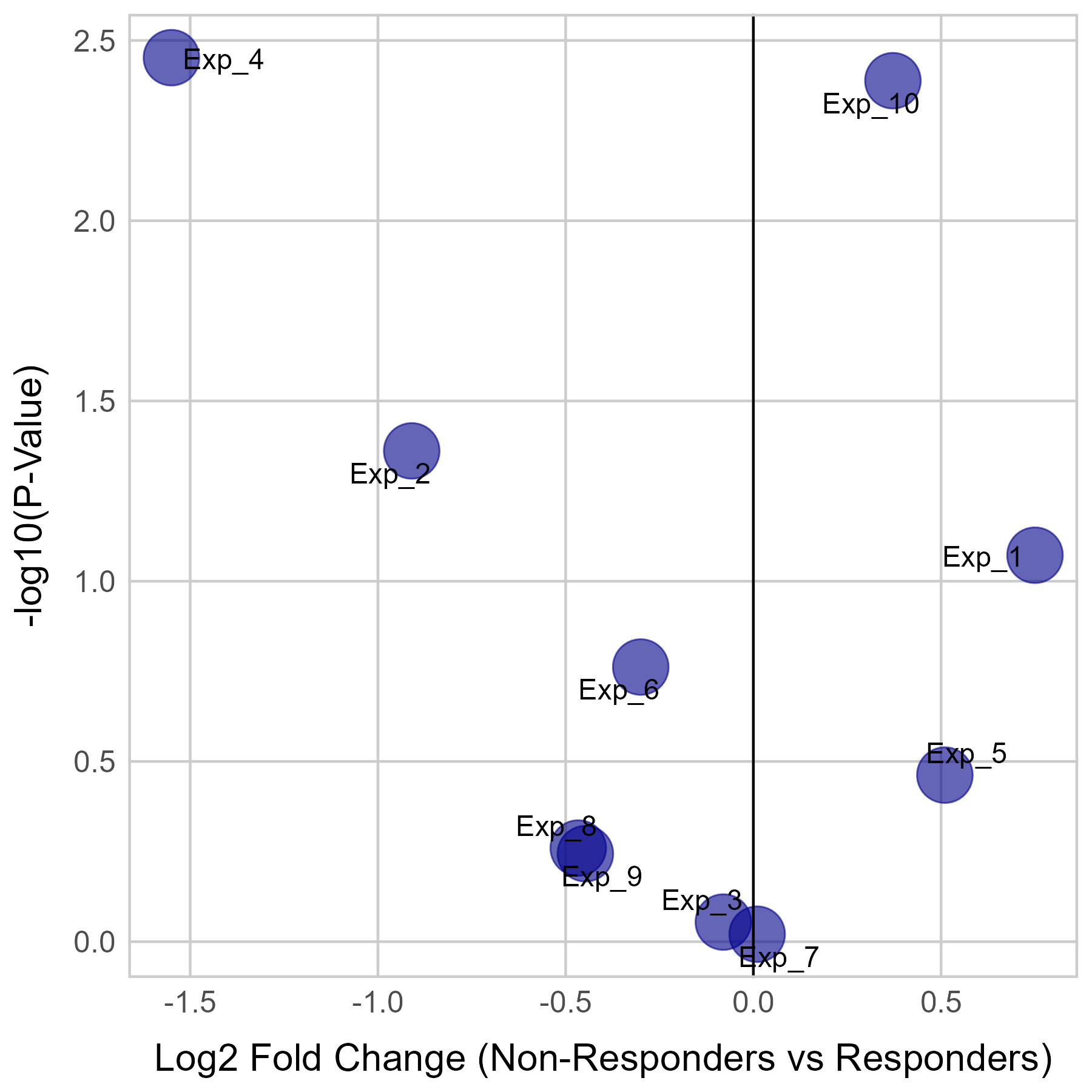 |  |
No significant differences were found in ICAM2 mutation.
Top |
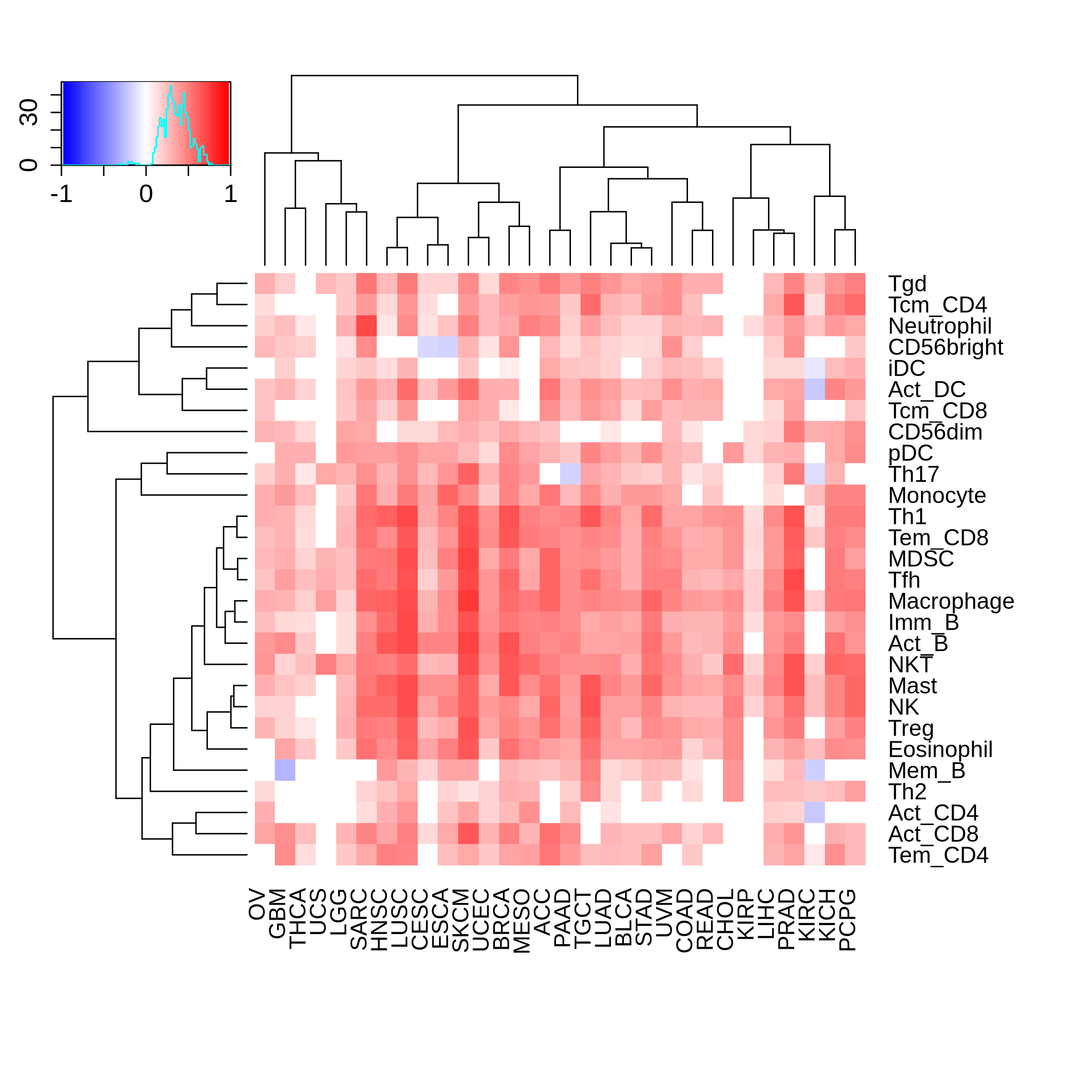
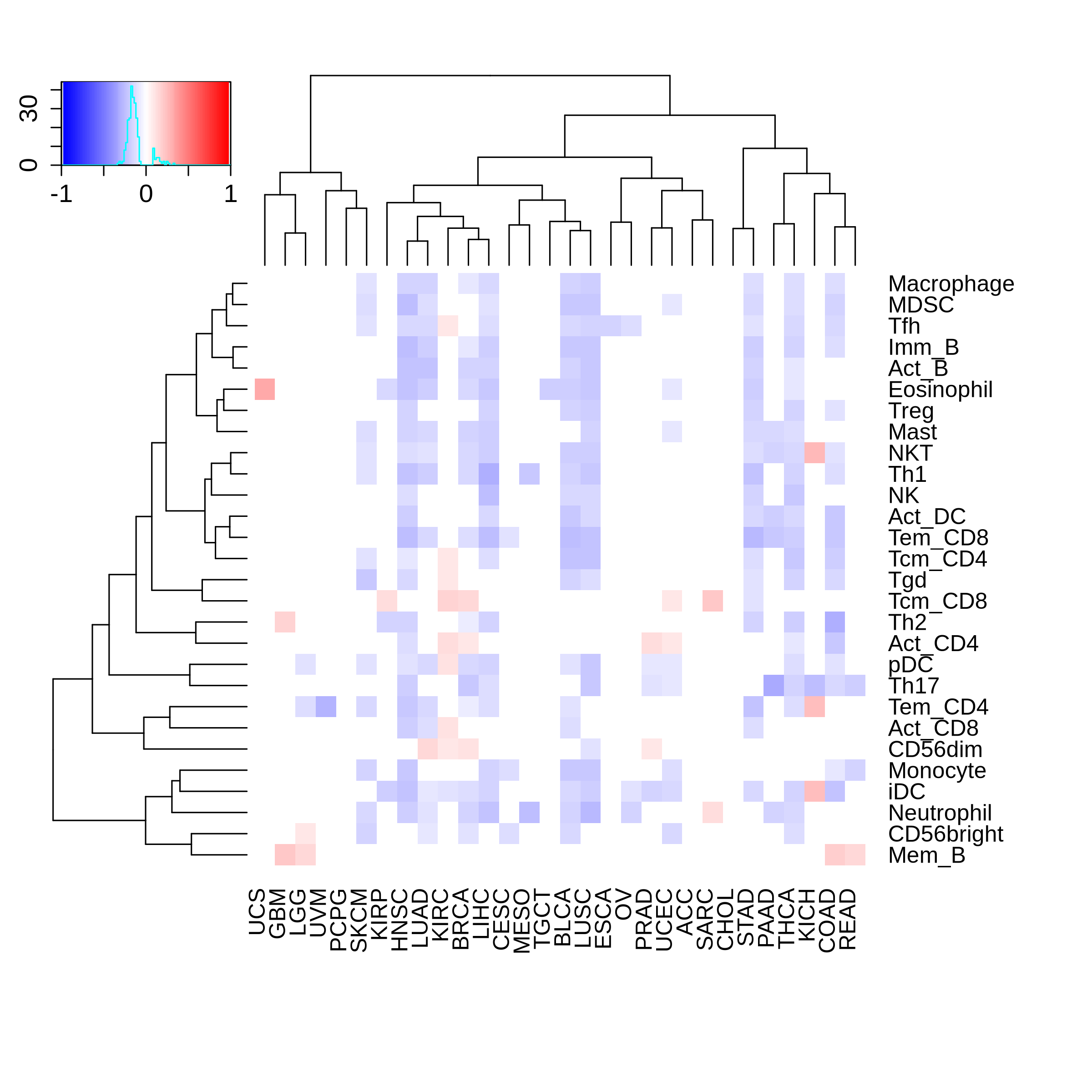
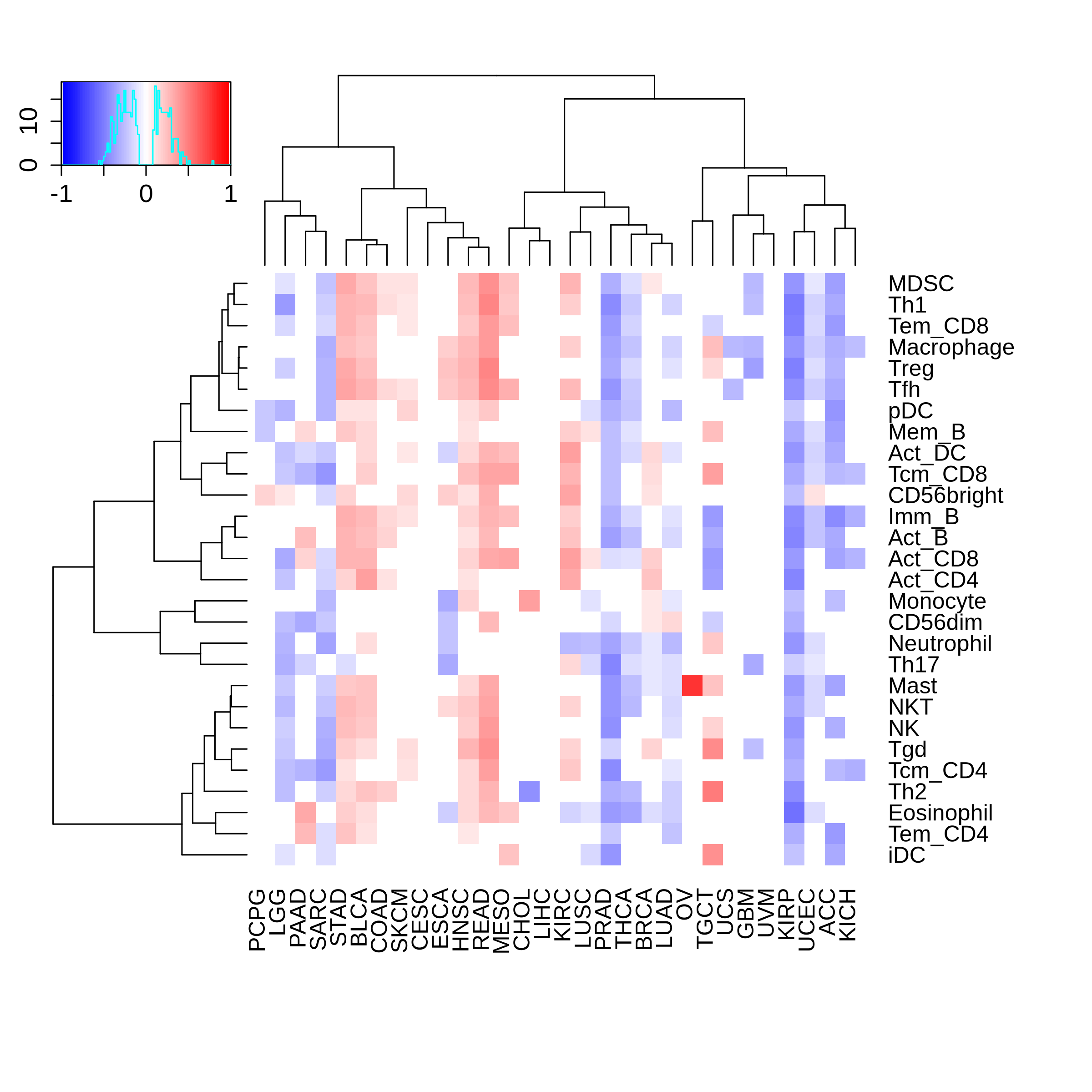
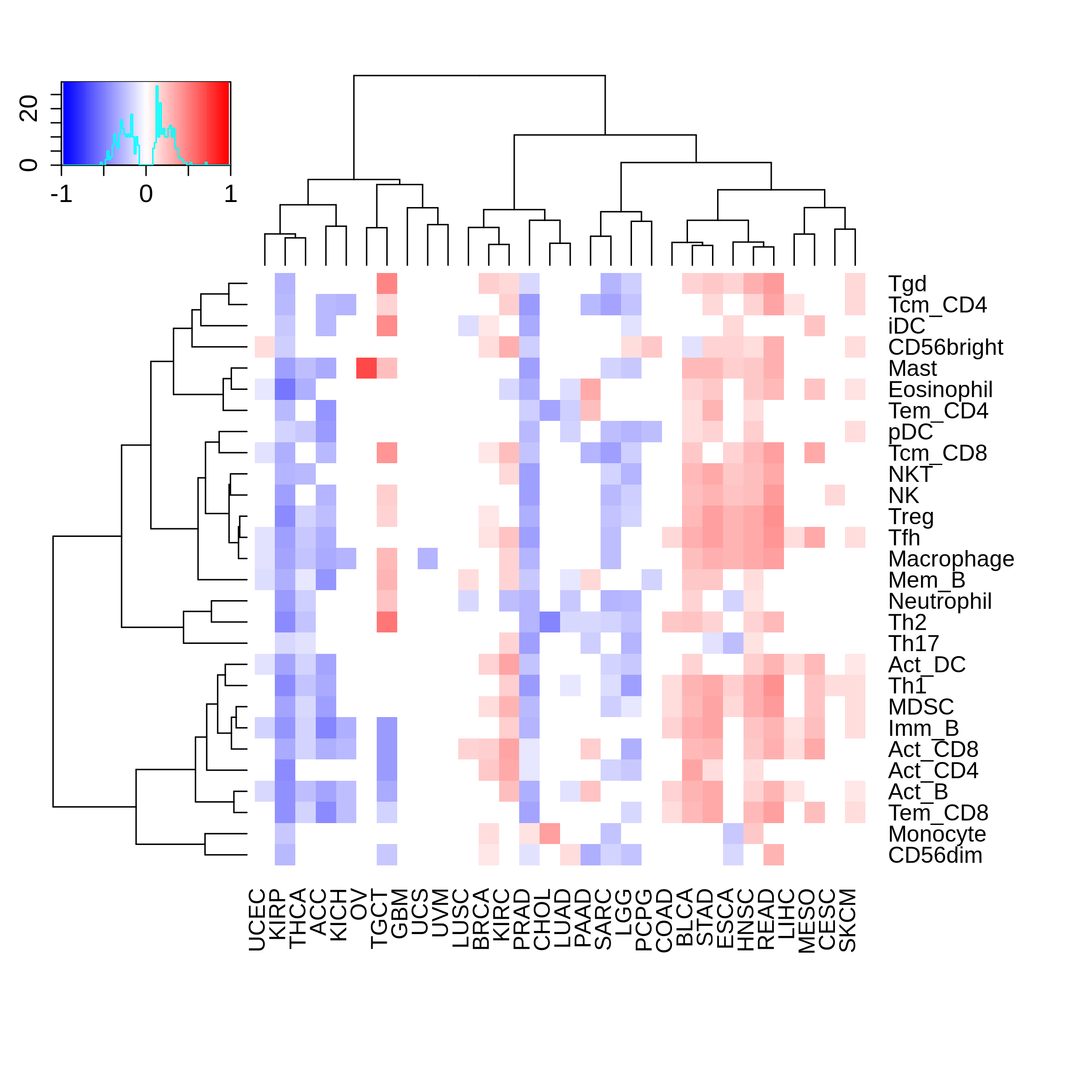
Correlation between the composition of TIL and gene expression, methylation and CNV |
Top |
The association between ICAM2 expression and immune subtypes/status |
Top |
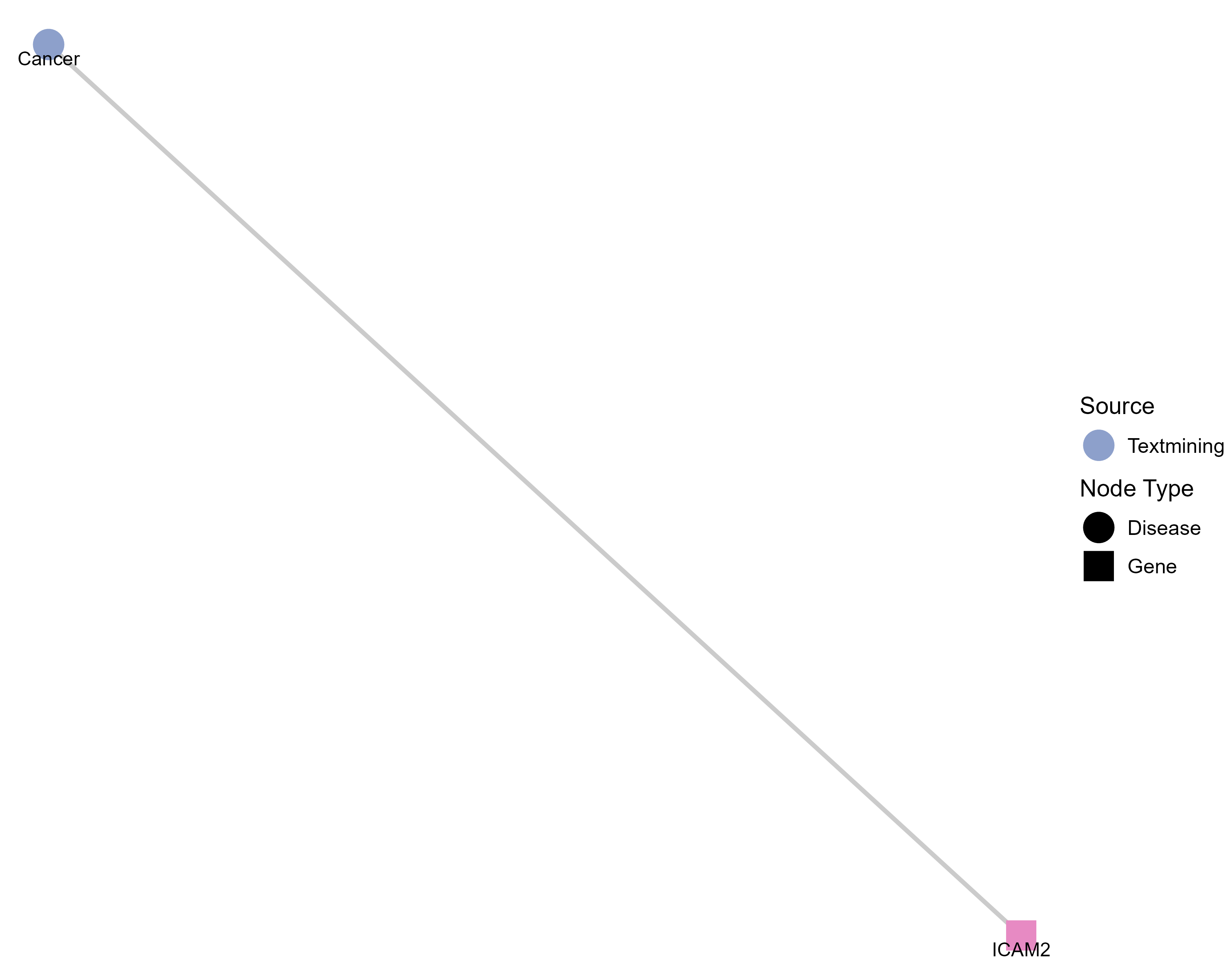
Drugs targeting ICAM2 and diseases related to ICAM2. |
Top |
Survival analysis based on ICAM2 expression |
Top |
| Reference |
| [1] Steele CD, Abbasi A, Islam SMA, et al. Signatures of copy number alterations in human cancer. Nature. 2022 Jun;606(7916):984-991. doi: 10.1038/s41586-022-04738-6. Epub 2022 Jun 15. PMID: 35705804; PMCID: PMC9242861. [2] Beibei Ru, Ching Ngar Wong, Yin Tong, et al. TISIDB: an integrated repository portal for tumor–immune system interactions, Bioinformatics, Volume 35, Issue 20, October 2019, Pages 4200–4202, https://doi.org/10.1093/bioinformatics/btz210. [3] Zhongyang Liu, Jiale Liu, Xinyue Liu, et al. CTR–DB, an omnibus for patient-derived gene expression signatures correlated with cancer drug response, Nucleic Acids Research, Volume 50, Issue D1, 7 January 2022, Pages D1184–D1199, https://doi.org/10.1093/nar/gkab860. [4] Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017 Jan 3;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. PMID: 28052254. [5] Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity. 2018 Apr 17;48(4):812-830.e14. doi: 10.1016/j.immuni.2018.03.023. Epub 2018 Apr 5. Erratum in: Immunity. 2019 Aug 20;51(2):411-412. doi: 10.1016/j.immuni.2019.08.004. PMID: 29628290; PMCID: PMC5982584. [6] Zapata L, Caravagna G, Williams MJ, et al. Immune selection determines tumor antigenicity and influences response to checkpoint inhibitors. Nat Genet. 2023 Mar;55(3):451-460. doi: 10.1038/s41588-023-01313-1. Epub 2023 Mar 9. PMID: 36894710; PMCID: PMC10011129. [7] Cortes-Ciriano I, Lee S, Park WY, et al. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017 Jun 6;8:15180. doi: 10.1038/ncomms15180. PMID: 28585546; PMCID: PMC5467167. [8] Cannon M, Stevenson J, Stahl K, et al. DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res. 2024 Jan 5;52(D1):D1227-D1235. doi: 10.1093/nar/gkad1040. PMID: 37953380; PMCID: PMC10767982. [9] Grissa D, Junge A, Oprea TI, Jensen LJ. Diseases 2.0: a weekly updated database of disease-gene associations from text mining and data integration. Database (Oxford). 2022 Mar 28;2022:baac019. doi: 10.1093/database/baac019. PMID: 35348648; PMCID: PMC9216524. |