
|
||||||
|
||||||
Navigation1. Fusion gene data collection 2. Protean features (domains) retention analysis 3. Creation of fusion transcript and amino acid sequences 4. Understanding of FusionGDB's annotation category Search page, example: ABL1 involved fusion genes (specifically, BCR-ABL1) FusionGene search result page FusionGene annotation result page 1) FusionGeneSummary category 2) FusionProtFeatures category 3) FusionGeneSequence category 4) FusionGenePPI 5) RelatedDrugs category 6) RelatedDiseases category 5. Download data and contact us 1. Fusion gene data collection We obtained fusion genes which are validated or predicted from Entrez mRNA sequence libraries and human primary tumor samples of TCGA data. The former was mainly downloaded from the most recent version of ChiTaRS-3.1 and the latter was obtained from two papers (Xin et al (2018) and Qingsong et al. (2018)). We adopted their gene list for each corresponding samples and we have 1,005 Sanger fusion genes and 30,270 TCGA fusion genes, respectively. All genome coordinates of break points were lifted over to hg19. Detailed information is in statistics page. 2. Protean features (domains) retention analysis We searched the retention of 39 protein features of UniProt (six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features) at the fusion amino acid sequence level. Through this process, we also checked the retention of protein-protein interaction (PPI) at the fusion protein. Detailed information about all of the protein features is in UniProt page. * Click on the image to enlarge it in a new window.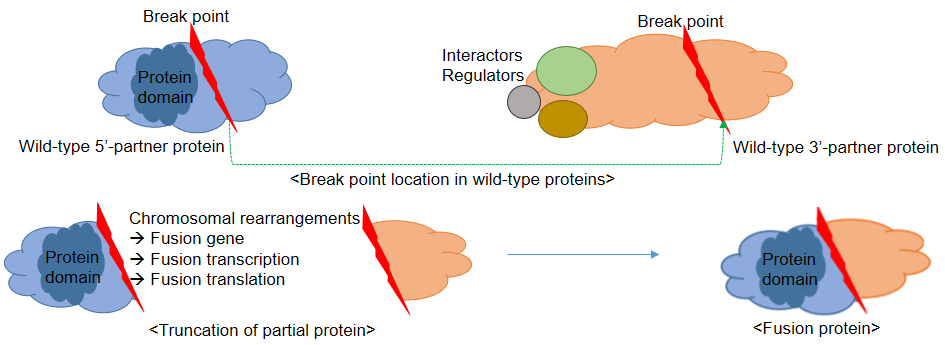 3. Creation of fusion transcript and amino acid sequences To help experimental research laboratories, we have created the fusion transcript sequence and fusion amino acid sequences of ~ 10K in-frame fusion genes. Here, we considered the matched isoforms for each gene. The users can download the fusion isoform sequences from each annotation page. 4. Understanding FusionGDB's annotation categories Search page, example: ABL1 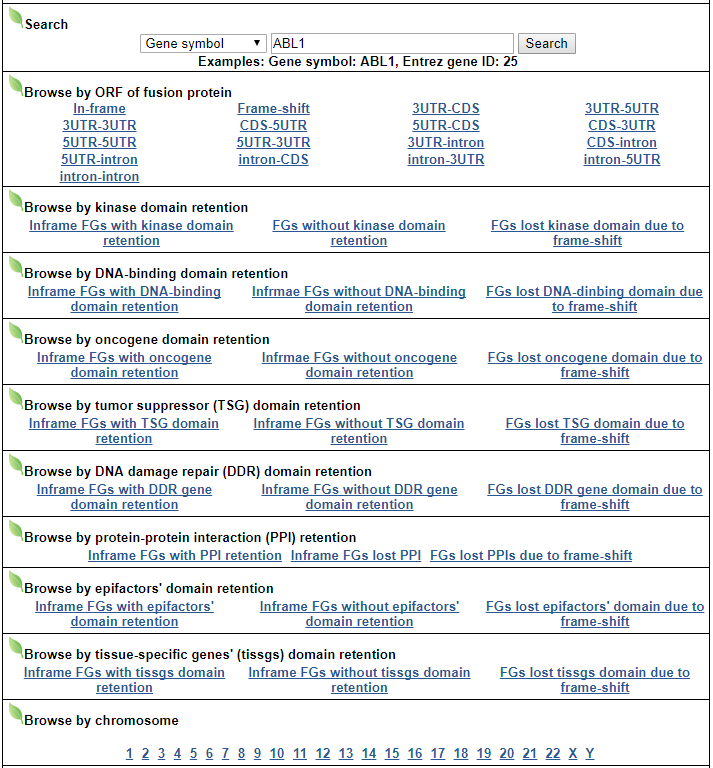 Input query - Official HUGO gene symbol or Entrez gene ID. FusionGene search result page 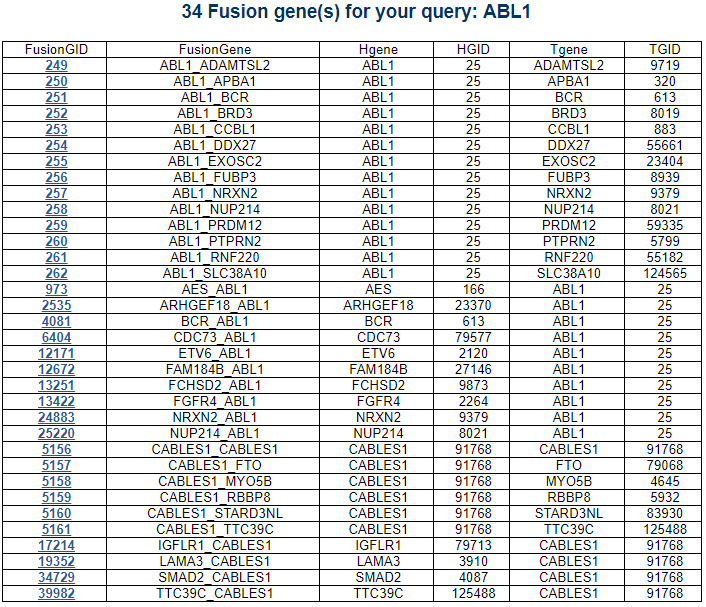 Select your fusion gene from the gene list. FusionGene annotation result page>  These are FusionGeneDB's annotation categories for your query with links to their corresponding annotation parts. 1) FusionGeneSummary category This category shows the information of fusion gene. Firstly, it shows each partner gene's overall information from basic information such as symbol, alias, and locations and ENST accessions involved in fusion gene. Specifically, DoF score provides the all possible combinations of each gene in pan-cancer fusion genes. From the # samples, the user can Words in blue are linked to their respective databases. FusionGeneSummary table also shows the tissue and cancer type information including manually curated PubMed article information. 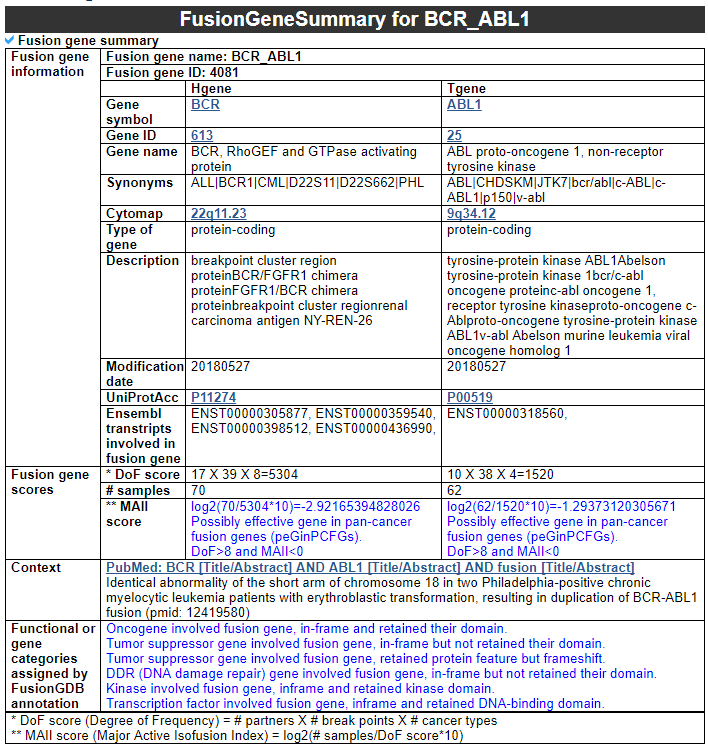 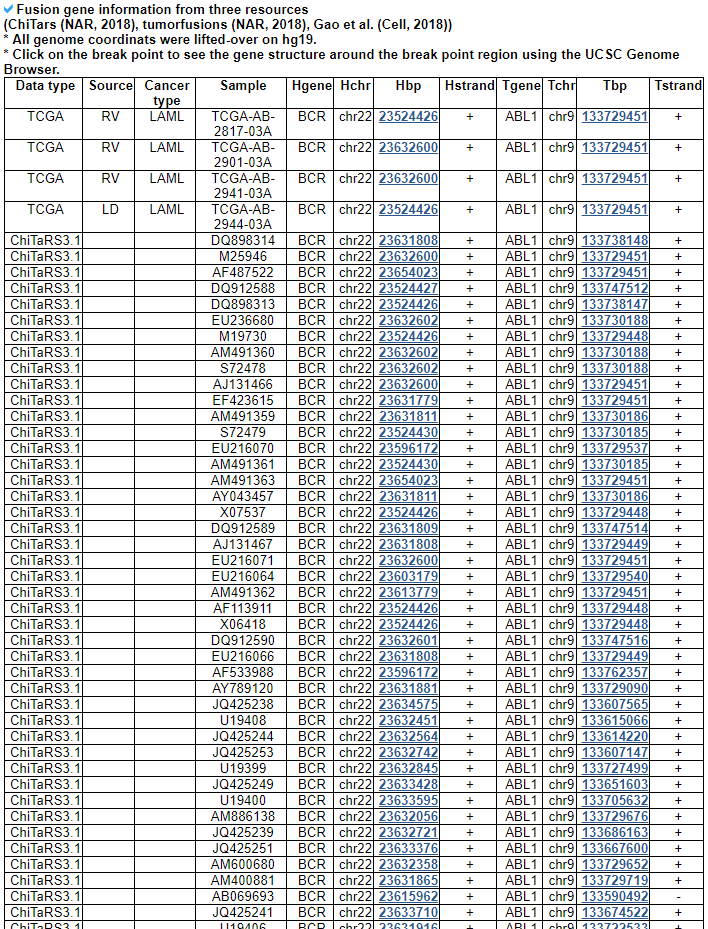 2) FusionProtFeatures category This category provides the protein domain retention information.  --- This image shows the protein function information of each fusion partner protein.  Retention analysis result of each fusion partner protein across 39 protein features of UniProt. (Six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features) Retention analysis result of each fusion partner protein across 39 protein features of UniProt. (Six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features)Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at download page .* Minus value of BPloci means that the break pointn is located before the CDS. - In-frame and retained protein feature among the 13 regional features. After fusion protein formed, these protein domains or features were still intact. 
 --- This image shows the retained protein features among 13 region features such as Topological domain, Transmembrane, Intramembrane, Domain, Repeat, Calcium binding, Zinc finger, DNA binding, Nucleotide binding, Region, Coiled coil, Motif, and Compositional bias in each fusion partner protein. - In-frame and not-retained protein feature among the 13 regional features. Due to the truncation by gene fusion, these protein domains or features were lost. 
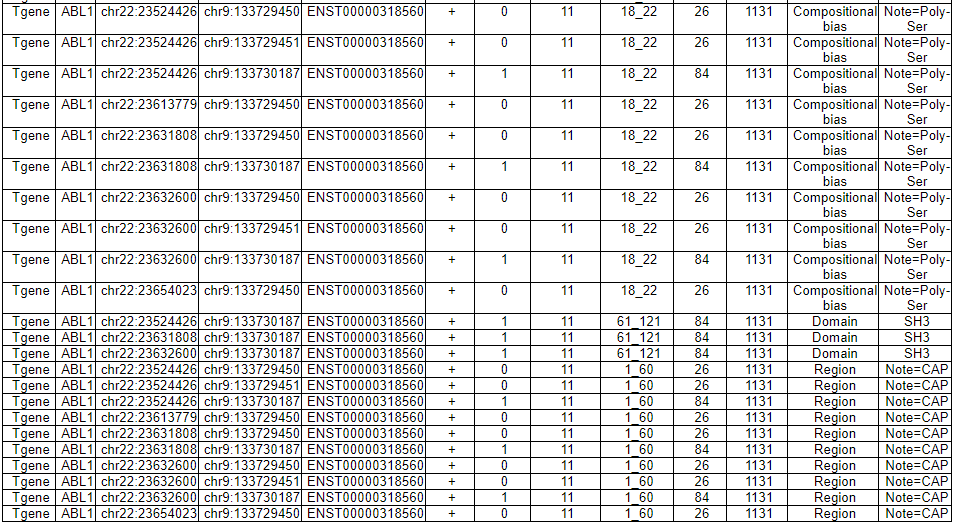 --- This image shows the not-retained protein features among 13 region features. 3) FusionGeneSequence category --- This category prvides the fusion transcript and amino acid sequences based on multiple iso-fusion gene structures.  * Fusion amino acid sequences.  * Fusion transcript sequences (CSD region only).  * Fusion transcript sequences (whole transcript).  4) FusionGenePPI category --- This image shows the protein-protein interactors and possible network link. 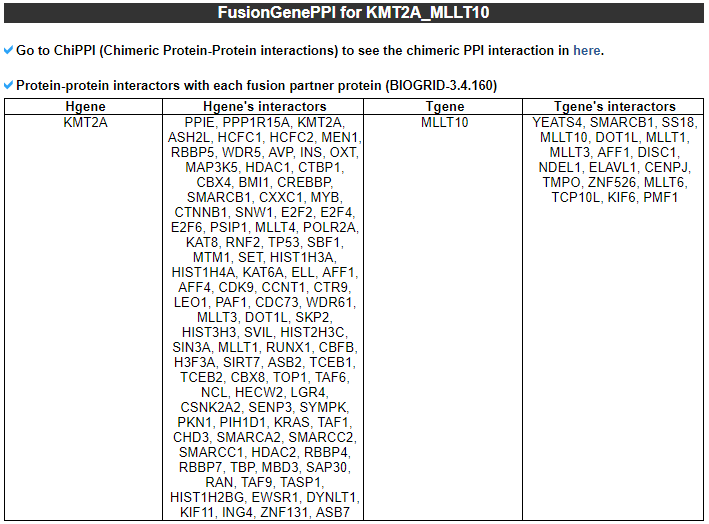 - In-frame with interaction retention. After fusion protein formed, the PPI still kept.  - In-frame without interaction retention. Due to fusion formation, the PPI has been lost. 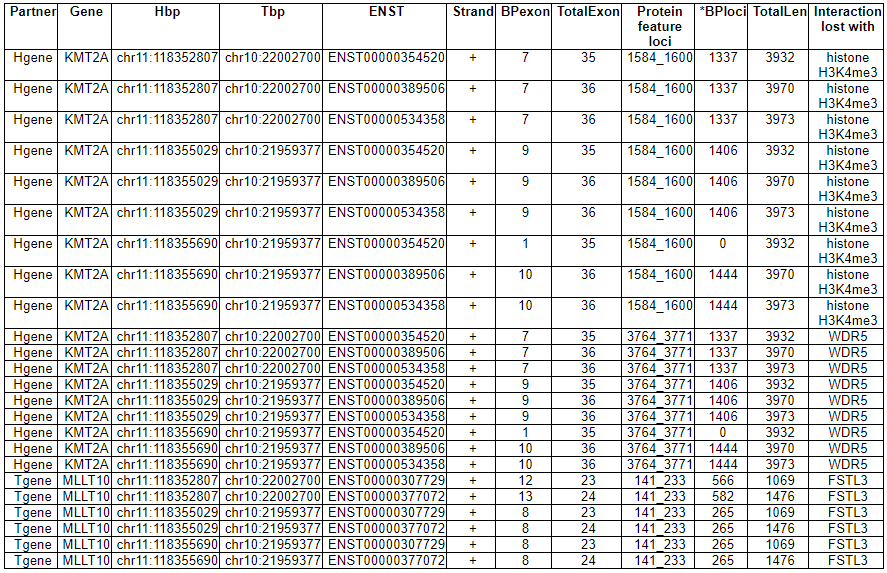 5) RelatedDrugs category --- This table provides the DrugBank information related with each fusion partner. 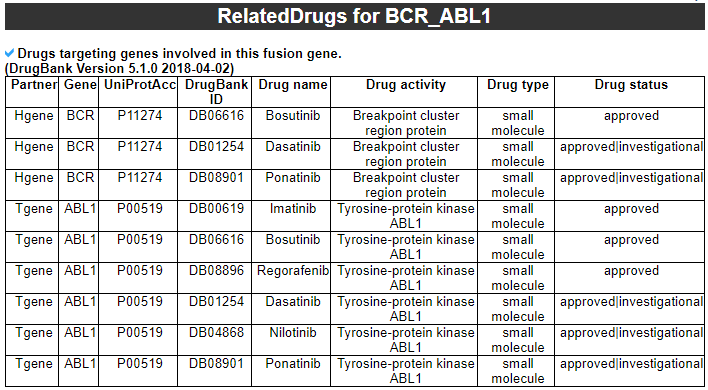
6) RelatedDiseases category --- This table provides disease information related with each fusion partner. 
5. Download data and contact us Please go to download page andcontact page . |
||||||
