
|
Home |
Download |
Statistics |
Landscape |
Help |
Contact |
Statistics and landscape
 Overview of ExonSkipDB
Overview of ExonSkipDB- 14,272 genes (14,223 UniProt Accessions) involved in 90,520 exon skipping events.
- We identified exon-skipping events that have four splice sites overlap among three exons involved in exon skipping event with GENCODE GRCh37 gene structure.
- Of these, 8,667 genes have in-frame exon skipping events.
- On the other hand, there were 9,623 frame-shift exon skipping events. These genes need further studies to be used as the antisense oligonucleotide targets.
 Lost protein functional features due to in-frame exon skipping events in TCGA.
Lost protein functional features due to in-frame exon skipping events in TCGA.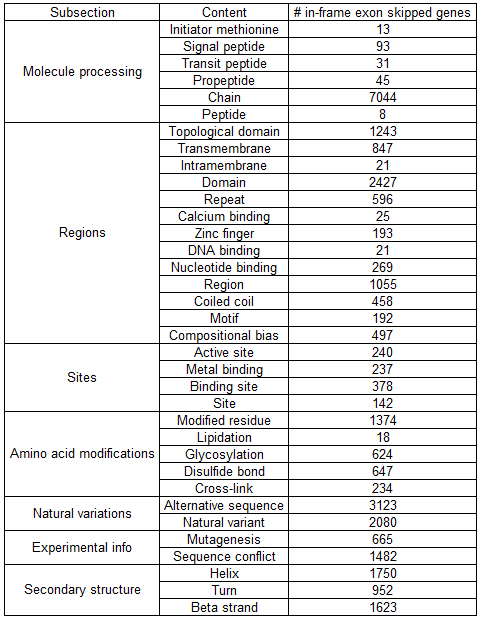
 249 genes (TFs) that have lost DNA-binding domain due to exon skipping events.
249 genes (TFs) that have lost DNA-binding domain due to exon skipping events.AEBP2, AMFR, ANKIB1, AR, ASH1L, BAZ2A, BCL6, BRCA1, CAMTA1, CAMTA2, CBLB, CBLC, CHD3, CHN1, CPSF4, CPSF4L, CREBBP, CTCF, CUL9, CUX2, CYHR1, DEF8, DEK, DGKA, DGKG, DNAJA1, DNAJC21, DNAJC24, DPF2, DTX1, DTX3L, E2F7, E2F8, EBF1, EBF3, EEA1, ELF1, ELK3, ESCO2, ESR1, ESRRA, ESRRB, ETV5, FGD2, FGD4, FLYWCH1, FOXJ2, FOXJ3, FOXK2, FOXM1, FOXP4, G2E3, GFI1B, GLI1, GLI3, GPATCH8, GTF2E2, HDAC6, HDX, HESX1, HHEX, HIVEP1, HMBOX1, HMG20B, HMGA1, HMGA2, IKZF1, IRF7, KAT7, KCMF1, KDM5B, KDM5C, KDM5D, KLF5, KMT2B, KMT2C, L3MBTL3, L3MBTL4, LEF1, LHX8, LIG3, MAPK1, MBNL1, MBNL2, MEF2C, MEF2D, MKRN2, MNAT1, MYBL1, MYBL2, MYRFL, MYT1, MYT1L, NABP2, NBR1, NFIA, NFX1, NFYA, NR1H2, NR1H3, NR1H4, NR3C1, NRF1, NSD1, NSMCE1, NUP153, PARP2, PATZ1, PDS5B, PGR, PHF1, PHF10, PHF14, PHF19, PHF21A, PMS1, POLR2K, POLR3B, POU2F3, PRDM1, PRDM4, PRDM5, PRKD2, RACGAP1, RBCK1, RBM10, RBM27, RBPJL, RERE, REST, RFX2, RFX5, RNF121, RNF13, RNF138, RNF144B, RNF167, RNF175, RNF181, RNF2, RNF216, RNF220, RNF25, RNF34, RNF8, RNFT1, RPA1, RPA2, RPS27, RSF1, RXRG, SAP30, SF3A3, SMYD2, SMYD3, SOBP, SOX30, SOX6, SPDEF, SPEN, SREK1IP1, TBX18, TBX2, TBX20, TBX3, TBX4, TCEA1, TCEA2, TCF7L1, TFAM, THAP7, TOX4, TRAF2, TRAF3, TRAF4, TRAF5, TRAF6, TRAF7, TRERF1, TRIM69, TRIP4, U2AF1, UBR1, UBR2, UBTF, UNK, USP3, USP33, VPS41, WBP4, WDFY1, YAF2, YY1, ZBTB11, ZBTB16, ZBTB17, ZBTB20, ZBTB40, ZBTB48, ZC3H11A, ZC3H8, ZCCHC10, ZCCHC24, ZCCHC9, ZFAND2A, ZFAND2B, ZFAND3, ZFAND5, ZFAND6, ZFHX3, ZFP64, ZFPM1, ZFPM2, ZFY, ZMAT3, ZMAT4, ZMAT5, ZMIZ1, ZMYM1, ZMYM4, ZMYND15, ZNF143, ZNF217, ZNF236, ZNF280C, ZNF318, ZNF330, ZNF346, ZNF384, ZNF385A, ZNF385D, ZNF407, ZNF410, ZNF423, ZNF462, ZNF512, ZNF512B, ZNF521, ZNF532, ZNF536, ZNF683, ZNF710, ZNF76, ZNF827, ZNF862, ZNHIT6, ZNRF3, ZSWIM7, and ZXDC.
 Overrepresentation Enrichment Analysis (ORA) of ES genes with recurrent mutations in the skipped exons per cancer type
Overrepresentation Enrichment Analysis (ORA) of ES genes with recurrent mutations in the skipped exons per cancer type
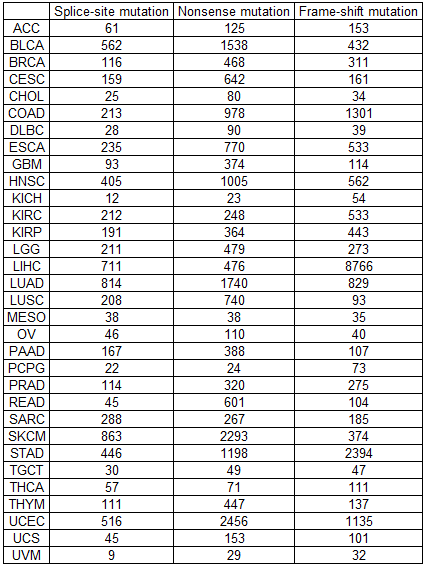
 Mutations in the skipped exons
Mutations in the skipped exons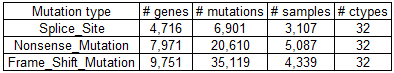

 t-distributed stochastic neighbor embedding (tSNE) of PSI values of sQTMs
t-distributed stochastic neighbor embedding (tSNE) of PSI values of sQTMs* Click on the image to enlarge it in a new window.

 tSNE of CG regions of sQTMs
tSNE of CG regions of sQTMs* Click on the image to enlarge it in a new window.

