| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_76491 | 11 | 85670079:85670103:85671717:85671820:85685750:85685855 | 85671717:85671820 | ENSG00000073921.13 | ENST00000530692.1 |
| exon_skip_76494 | 11 | 85670079:85670103:85685750:85685855:85687665:85687725 | 85685750:85685855 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526961.1,ENST00000526548.1,ENST00000526033.1,ENST00000528398.1,ENST00000529760.1,ENST00000393346.3 |
| exon_skip_76495 | 11 | 85670079:85670103:85685750:85685855:85692171:85692271 | 85685750:85685855 | ENSG00000073921.13 | ENST00000356360.5,ENST00000532603.1 |
| exon_skip_76498 | 11 | 85670079:85670103:85692171:85692271:85692787:85692818 | 85692171:85692271 | ENSG00000073921.13 | ENST00000529016.1 |
| exon_skip_76500 | 11 | 85685824:85685855:85687665:85687725:85689112:85689136 | 85687665:85687725 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526907.1,ENST00000526961.1,ENST00000529760.1,ENST00000530692.1 |
| exon_skip_76501 | 11 | 85685824:85685855:85687665:85687725:85692171:85692271 | 85687665:85687725 | ENSG00000073921.13 | ENST00000526033.1,ENST00000528398.1,ENST00000393346.3 |
| exon_skip_76504 | 11 | 85685824:85685855:85692171:85692271:85692787:85692818 | 85692171:85692271 | ENSG00000073921.13 | ENST00000356360.5,ENST00000532603.1 |
| exon_skip_76505 | 11 | 85687691:85687725:85689112:85689136:85692171:85692271 | 85689112:85689136 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526961.1,ENST00000529760.1,ENST00000530692.1 |
| exon_skip_76509 | 11 | 85692914:85693031:85694908:85695016:85707868:85707972 | 85694908:85695016 | ENSG00000073921.13 | ENST00000529760.1 |
| exon_skip_76512 | 11 | 85692914:85693046:85694908:85695016:85701292:85701301 | 85694908:85695016 | ENSG00000073921.13 | ENST00000356360.5,ENST00000526033.1,ENST00000393346.3 |
| exon_skip_76513 | 11 | 85692914:85693046:85694908:85695016:85707868:85707972 | 85694908:85695016 | ENSG00000073921.13 | ENST00000532317.1,ENST00000529016.1,ENST00000528398.1 |
| exon_skip_76519 | 11 | 85694908:85695016:85701292:85701421:85707868:85707972 | 85701292:85701421 | ENSG00000073921.13 | ENST00000526961.1,ENST00000526033.1 |
| exon_skip_76520 | 11 | 85694908:85695016:85701292:85701442:85707868:85707972 | 85701292:85701442 | ENSG00000073921.13 | ENST00000530542.1,ENST00000356360.5,ENST00000393346.3 |
| exon_skip_76523 | 11 | 85707868:85707972:85711685:85711822:85712077:85712184 | 85711685:85711822 | ENSG00000073921.13 | ENST00000532317.1,ENST00000530542.1,ENST00000529016.1,ENST00000356360.5,ENST00000526033.1,ENST00000528398.1,ENST00000529760.1,ENST00000393346.3 |
| exon_skip_76525 | 11 | 85714408:85714494:85718584:85718626:85722072:85722178 | 85718584:85718626 | ENSG00000073921.13 | ENST00000532317.1,ENST00000356360.5,ENST00000534412.1,ENST00000526033.1,ENST00000528398.1,ENST00000393346.3 |
| exon_skip_76526 | 11 | 85722154:85722179:85723323:85723435:85725912:85726006 | 85723323:85723435 | ENSG00000073921.13 | ENST00000532317.1,ENST00000531771.1,ENST00000356360.5,ENST00000531558.1,ENST00000534412.1,ENST00000526033.1,ENST00000528398.1,ENST00000393346.3 |
| exon_skip_76527 | 11 | 85725912:85726006:85733409:85733512:85737333:85737409 | 85733409:85733512 | ENSG00000073921.13 | ENST00000532317.1,ENST00000525162.1,ENST00000356360.5,ENST00000528256.1,ENST00000526033.1,ENST00000528398.1,ENST00000531930.1,ENST00000393346.3 |
| exon_skip_76528 | 11 | 85725912:85726006:85733409:85733512:85742510:85742653 | 85733409:85733512 | ENSG00000073921.13 | ENST00000532041.1 |
| exon_skip_76533 | 11 | 85733420:85733512:85734458:85734584:85737333:85737409 | 85734458:85734584 | ENSG00000073921.13 | ENST00000528411.1 |
| exon_skip_76534 | 11 | 85733420:85733512:85737333:85737409:85742510:85742653 | 85737333:85737409 | ENSG00000073921.13 | ENST00000532317.1,ENST00000525162.1,ENST00000356360.5,ENST00000528256.1,ENST00000526033.1,ENST00000528398.1,ENST00000531930.1,ENST00000393346.3 |
| exon_skip_76536 | 11 | 85733420:85733512:85742510:85742653:85779692:85779775 | 85742510:85742653 | ENSG00000073921.13 | ENST00000532041.1 |
| exon_skip_76537 | 11 | 85737333:85737409:85742510:85742653:85779692:85779775 | 85742510:85742653 | ENSG00000073921.13 | ENST00000356360.5,ENST00000528256.1,ENST00000526033.1,ENST00000528411.1,ENST00000531930.1,ENST00000393346.3 |
| exon_skip_76539 | 11 | 85742510:85742653:85779692:85779775:85780761:85780853 | 85779692:85779775 | ENSG00000073921.13 | ENST00000531930.1 |
| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_76491 | 11 | 85670079:85670103:85671717:85671820:85685750:85685855 | 85671717:85671820 | ENSG00000073921.13 | ENST00000530692.1 |
| exon_skip_76494 | 11 | 85670079:85670103:85685750:85685855:85687665:85687725 | 85685750:85685855 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526033.1,ENST00000526548.1,ENST00000393346.3,ENST00000529760.1,ENST00000528398.1,ENST00000526961.1 |
| exon_skip_76495 | 11 | 85670079:85670103:85685750:85685855:85692171:85692271 | 85685750:85685855 | ENSG00000073921.13 | ENST00000532603.1,ENST00000356360.5 |
| exon_skip_76498 | 11 | 85670079:85670103:85692171:85692271:85692787:85692818 | 85692171:85692271 | ENSG00000073921.13 | ENST00000529016.1 |
| exon_skip_76500 | 11 | 85685824:85685855:85687665:85687725:85689112:85689136 | 85687665:85687725 | ENSG00000073921.13 | ENST00000532317.1,ENST00000530692.1,ENST00000529760.1,ENST00000526961.1,ENST00000526907.1 |
| exon_skip_76501 | 11 | 85685824:85685855:85687665:85687725:85692171:85692271 | 85687665:85687725 | ENSG00000073921.13 | ENST00000526033.1,ENST00000393346.3,ENST00000528398.1 |
| exon_skip_76504 | 11 | 85685824:85685855:85692171:85692271:85692787:85692818 | 85692171:85692271 | ENSG00000073921.13 | ENST00000532603.1,ENST00000356360.5 |
| exon_skip_76505 | 11 | 85687691:85687725:85689112:85689136:85692171:85692271 | 85689112:85689136 | ENSG00000073921.13 | ENST00000532317.1,ENST00000530692.1,ENST00000529760.1,ENST00000526961.1 |
| exon_skip_76509 | 11 | 85692914:85693031:85694908:85695016:85707868:85707972 | 85694908:85695016 | ENSG00000073921.13 | ENST00000529760.1 |
| exon_skip_76512 | 11 | 85692914:85693046:85694908:85695016:85701292:85701301 | 85694908:85695016 | ENSG00000073921.13 | ENST00000526033.1,ENST00000393346.3,ENST00000356360.5 |
| exon_skip_76513 | 11 | 85692914:85693046:85694908:85695016:85707868:85707972 | 85694908:85695016 | ENSG00000073921.13 | ENST00000532317.1,ENST00000528398.1,ENST00000529016.1 |
| exon_skip_76519 | 11 | 85694908:85695016:85701292:85701421:85707868:85707972 | 85701292:85701421 | ENSG00000073921.13 | ENST00000526033.1,ENST00000526961.1 |
| exon_skip_76520 | 11 | 85694908:85695016:85701292:85701442:85707868:85707972 | 85701292:85701442 | ENSG00000073921.13 | ENST00000393346.3,ENST00000356360.5,ENST00000530542.1 |
| exon_skip_76523 | 11 | 85707868:85707972:85711685:85711822:85712077:85712184 | 85711685:85711822 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526033.1,ENST00000393346.3,ENST00000529760.1,ENST00000528398.1,ENST00000356360.5,ENST00000529016.1,ENST00000530542.1 |
| exon_skip_76525 | 11 | 85714408:85714494:85718584:85718626:85722072:85722178 | 85718584:85718626 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526033.1,ENST00000393346.3,ENST00000528398.1,ENST00000356360.5,ENST00000534412.1 |
| exon_skip_76526 | 11 | 85722154:85722179:85723323:85723435:85725912:85726006 | 85723323:85723435 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526033.1,ENST00000393346.3,ENST00000528398.1,ENST00000356360.5,ENST00000534412.1,ENST00000531771.1,ENST00000531558.1 |
| exon_skip_76527 | 11 | 85725912:85726006:85733409:85733512:85737333:85737409 | 85733409:85733512 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526033.1,ENST00000393346.3,ENST00000528398.1,ENST00000356360.5,ENST00000531930.1,ENST00000525162.1,ENST00000528256.1 |
| exon_skip_76528 | 11 | 85725912:85726006:85733409:85733512:85742510:85742653 | 85733409:85733512 | ENSG00000073921.13 | ENST00000532041.1 |
| exon_skip_76533 | 11 | 85733420:85733512:85734458:85734584:85737333:85737409 | 85734458:85734584 | ENSG00000073921.13 | ENST00000528411.1 |
| exon_skip_76534 | 11 | 85733420:85733512:85737333:85737409:85742510:85742653 | 85737333:85737409 | ENSG00000073921.13 | ENST00000532317.1,ENST00000526033.1,ENST00000393346.3,ENST00000528398.1,ENST00000356360.5,ENST00000531930.1,ENST00000525162.1,ENST00000528256.1 |
| exon_skip_76536 | 11 | 85733420:85733512:85742510:85742653:85779692:85779775 | 85742510:85742653 | ENSG00000073921.13 | ENST00000532041.1 |
| exon_skip_76537 | 11 | 85737333:85737409:85742510:85742653:85779692:85779775 | 85742510:85742653 | ENSG00000073921.13 | ENST00000526033.1,ENST00000393346.3,ENST00000356360.5,ENST00000531930.1,ENST00000528256.1,ENST00000528411.1 |
| exon_skip_76539 | 11 | 85742510:85742653:85779692:85779775:85780761:85780853 | 85779692:85779775 | ENSG00000073921.13 | ENST00000531930.1 |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q13492 | 255 | 269 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 255 | 269 | 221 | 294 | Region | Note=Interaction with PIMREG;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:16491119;Dbxref=PMID:16491119 |
| Q13492 | 419 | 469 | 420 | 469 | Alternative sequence | ID=VSP_009607;Note=In isoform 3 and isoform 4. Missing;Ontology_term=ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:14702039,ECO:0000303|PubMed:15489334;Dbxref=PMID:14702039,PMID:15489334 |
| Q13492 | 419 | 469 | 420 | 426 | Alternative sequence | ID=VSP_044568;Note=In isoform 5. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|Ref.3 |
| Q13492 | 419 | 469 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 469 | 505 | 420 | 469 | Alternative sequence | ID=VSP_009607;Note=In isoform 3 and isoform 4. Missing;Ontology_term=ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:14702039,ECO:0000303|PubMed:15489334;Dbxref=PMID:14702039,PMID:15489334 |
| Q13492 | 469 | 505 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 593 | 613 | 593 | 593 | Alternative sequence | ID=VSP_009608;Note=In isoform 3. M->MNGMHFPQY;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:15489334;Dbxref=PMID:15489334 |
| Q13492 | 593 | 613 | 594 | 613 | Alternative sequence | ID=VSP_004067;Note=In isoform 2. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:9737689;Dbxref=PMID:9737689 |
| Q13492 | 593 | 613 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 613 | 648 | 594 | 613 | Alternative sequence | ID=VSP_004067;Note=In isoform 2. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:9737689;Dbxref=PMID:9737689 |
| Q13492 | 613 | 648 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 613 | 648 | 641 | 641 | Natural variant | ID=VAR_028195;Note=F->L;Dbxref=dbSNP:rs556337 |
| Q13492 | 613 | 648 | 648 | 649 | Site | Note=Breakpoint for translocation to form CALM/MLLT10 fusion protein |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q13492 | 255 | 269 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 255 | 269 | 221 | 294 | Region | Note=Interaction with PIMREG;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:16491119;Dbxref=PMID:16491119 |
| Q13492 | 419 | 469 | 420 | 469 | Alternative sequence | ID=VSP_009607;Note=In isoform 3 and isoform 4. Missing;Ontology_term=ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:14702039,ECO:0000303|PubMed:15489334;Dbxref=PMID:14702039,PMID:15489334 |
| Q13492 | 419 | 469 | 420 | 426 | Alternative sequence | ID=VSP_044568;Note=In isoform 5. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|Ref.3 |
| Q13492 | 419 | 469 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 469 | 505 | 420 | 469 | Alternative sequence | ID=VSP_009607;Note=In isoform 3 and isoform 4. Missing;Ontology_term=ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:14702039,ECO:0000303|PubMed:15489334;Dbxref=PMID:14702039,PMID:15489334 |
| Q13492 | 469 | 505 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 593 | 613 | 593 | 593 | Alternative sequence | ID=VSP_009608;Note=In isoform 3. M->MNGMHFPQY;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:15489334;Dbxref=PMID:15489334 |
| Q13492 | 593 | 613 | 594 | 613 | Alternative sequence | ID=VSP_004067;Note=In isoform 2. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:9737689;Dbxref=PMID:9737689 |
| Q13492 | 593 | 613 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 613 | 648 | 594 | 613 | Alternative sequence | ID=VSP_004067;Note=In isoform 2. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:9737689;Dbxref=PMID:9737689 |
| Q13492 | 613 | 648 | 2 | 652 | Chain | ID=PRO_0000187062;Note=Phosphatidylinositol-binding clathrin assembly protein |
| Q13492 | 613 | 648 | 641 | 641 | Natural variant | ID=VAR_028195;Note=F->L;Dbxref=dbSNP:rs556337 |
| Q13492 | 613 | 648 | 648 | 649 | Site | Note=Breakpoint for translocation to form CALM/MLLT10 fusion protein |
| Depth of coverage in three exons | Mutation description |
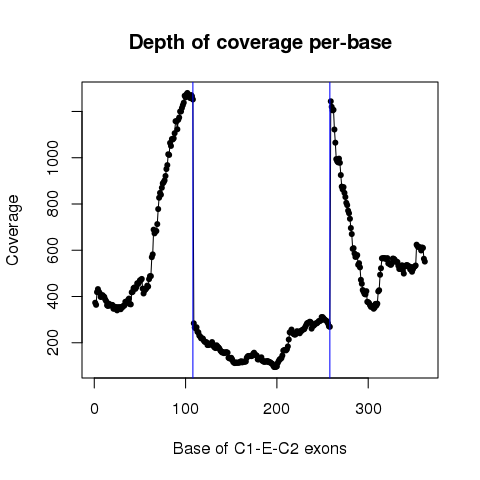 | Sample: TCGA-18-3410-01 |
| Cancer type: LUSC |
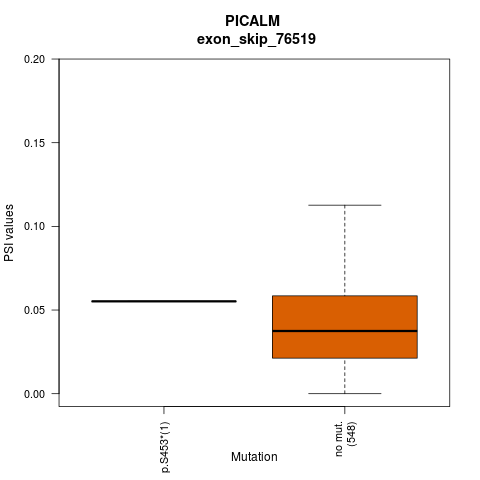
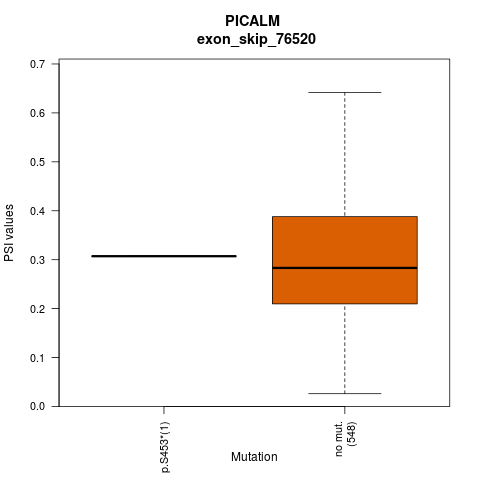
| ESID: exon_skip_76520 |
| Skipped exon start: 85701293 |
| Skipped exon end: 85701442 |
| Mutation start: 85701343 |
| Mutation end: 85701343 |
| Mutation type: Nonsense_Mutation |
| Reference seq: G |
| Mutation seq: C |
| AAchange: p.S453* |
 | Sample: TCGA-18-3410-01 |
| Cancer type: LUSC |
| ESID: exon_skip_76519 |
| Skipped exon start: 85701293 |
| Skipped exon end: 85701421 |
| Mutation start: 85701343 |
| Mutation end: 85701343 |
| Mutation type: Nonsense_Mutation |
| Reference seq: G |
| Mutation seq: C |
| AAchange: p.S453* |
exon_skip_76519_LUSC_TCGA-18-3410-01.png
 |
exon_skip_76520_LUSC_TCGA-18-3410-01.png
 |
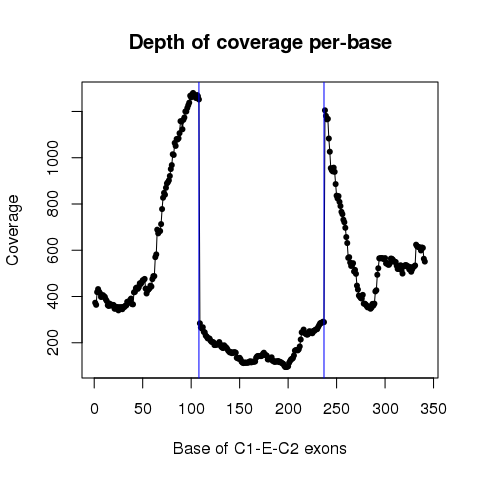 | Sample: TCGA-18-3410-01 |
| Cancer type: LUSC |
| ESID: exon_skip_76520 |
| Skipped exon start: 85701293 |
| Skipped exon end: 85701442 |
| Mutation start: 85701343 |
| Mutation end: 85701343 |
| Mutation type: Nonsense_Mutation |
| Reference seq: G |
| Mutation seq: C |
| AAchange: p.S453* |
 | Sample: TCGA-18-3410-01 |
| Cancer type: LUSC |
| ESID: exon_skip_76519 |
| Skipped exon start: 85701293 |
| Skipped exon end: 85701421 |
| Mutation start: 85701343 |
| Mutation end: 85701343 |
| Mutation type: Nonsense_Mutation |
| Reference seq: G |
| Mutation seq: C |
| AAchange: p.S453* |
exon_skip_76519_LUSC_TCGA-18-3410-01.png
 |
exon_skip_76520_LUSC_TCGA-18-3410-01.png
 |
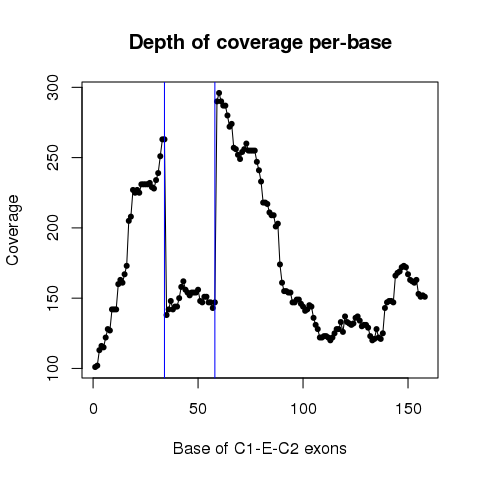 | Sample: TCGA-F4-6856-01 |
| Cancer type: COAD |
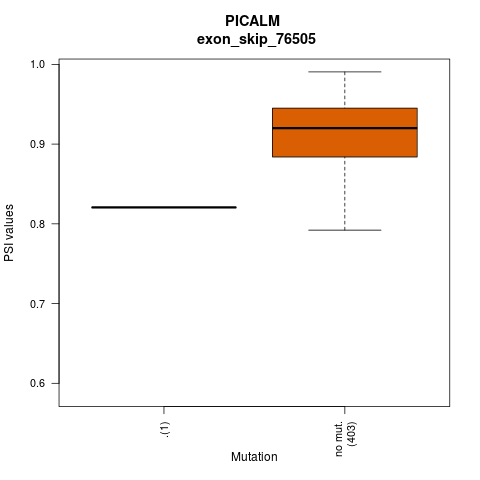
| ESID: exon_skip_76505 |
| Skipped exon start: 85689113 |
| Skipped exon end: 85689136 |
| Mutation start: 85689112 |
| Mutation end: 85689112 |
| Mutation type: Splice_Site |
| Reference seq: C |
| Mutation seq: T |
| AAchange: . |
exon_skip_318867_COAD_TCGA-F4-6856-01.png
 |
exon_skip_347508_COAD_TCGA-F4-6856-01.png
 |
exon_skip_449553_COAD_TCGA-F4-6856-01.png
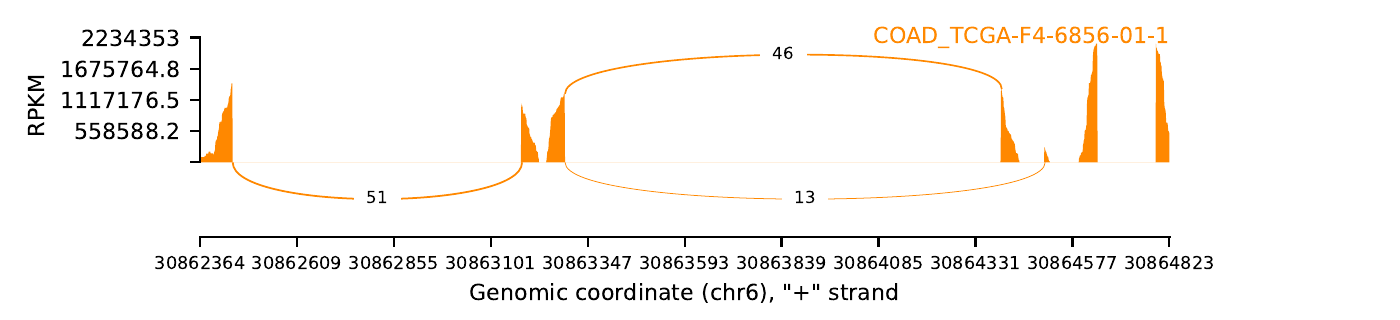 |
exon_skip_500368_COAD_TCGA-F4-6856-01.png
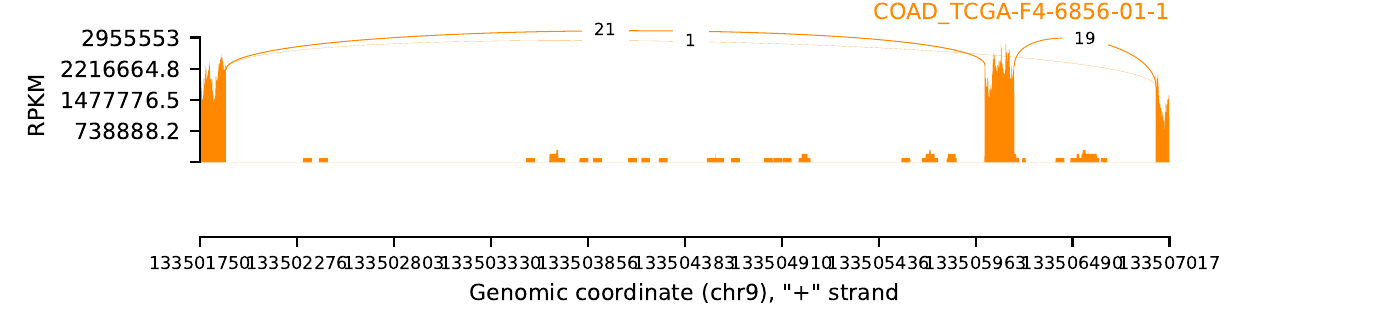 |
exon_skip_76505_COAD_TCGA-F4-6856-01.png
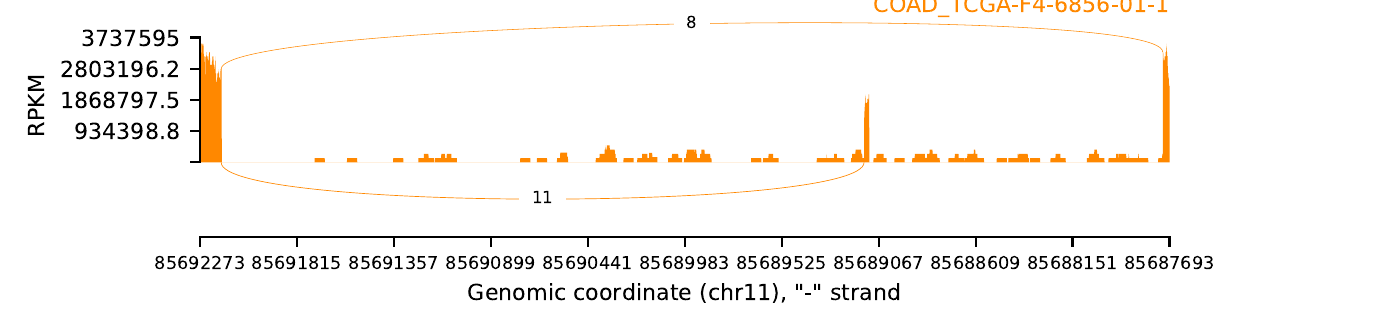 |
| Sample | Skipped exon start | Skipped exon end | Mutation start | Mutation end | Mutation type | Reference seq | Mutation seq | AAchange |
| SUPT1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85687666 | 85687725 | 85687718 | 85687718 | Frame_Shift_Del | G | - | p.P596fs |
| LN215_CENTRAL_NERVOUS_SYSTEM | 85718585 | 85718626 | 85718608 | 85718609 | Frame_Shift_Del | CT | - | p.G262fs |
| OC316_OVARY | 85737334 | 85737409 | 85737354 | 85737354 | Frame_Shift_Del | A | - | p.L110fs |
| RKO_LARGE_INTESTINE | 85685751 | 85685855 | 85685762 | 85685762 | Missense_Mutation | A | G | p.S645P |
| KYSE140_OESOPHAGUS | 85685751 | 85685855 | 85685813 | 85685813 | Missense_Mutation | A | C | p.L628V |
| CW2_LARGE_INTESTINE | 85692172 | 85692271 | 85692252 | 85692252 | Missense_Mutation | G | A | p.P567S |
| DAUDI_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85692172 | 85692271 | 85692263 | 85692263 | Missense_Mutation | T | C | p.N563S |
| BICR18_UPPER_AERODIGESTIVE_TRACT | 85692172 | 85692271 | 85692263 | 85692263 | Missense_Mutation | T | C | p.N563S |
| KYSE180_OESOPHAGUS | 85694909 | 85695016 | 85694971 | 85694971 | Missense_Mutation | A | G | p.L485P |
| DAUDI_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85694909 | 85695016 | 85694984 | 85694985 | Missense_Mutation | GG | AA | p.P481S |
| HCT15_LARGE_INTESTINE | 85701293 | 85701442 | 85701341 | 85701341 | Missense_Mutation | C | A | p.D454Y |
| HCT15_LARGE_INTESTINE | 85701293 | 85701421 | 85701341 | 85701341 | Missense_Mutation | C | A | p.D454Y |
| HRT18_LARGE_INTESTINE | 85701293 | 85701442 | 85701341 | 85701341 | Missense_Mutation | C | A | p.D454Y |
| HRT18_LARGE_INTESTINE | 85701293 | 85701421 | 85701341 | 85701341 | Missense_Mutation | C | A | p.D454Y |
| THP1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85701293 | 85701442 | 85701401 | 85701401 | Missense_Mutation | G | C | p.P434A |
| THP1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85701293 | 85701421 | 85701401 | 85701401 | Missense_Mutation | G | C | p.P434A |
| DAUDI_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85701293 | 85701442 | 85701411 | 85701411 | Missense_Mutation | A | C | p.D430E |
| DAUDI_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85701293 | 85701421 | 85701411 | 85701411 | Missense_Mutation | A | C | p.D430E |
| BICR18_UPPER_AERODIGESTIVE_TRACT | 85701293 | 85701442 | 85701411 | 85701411 | Missense_Mutation | A | C | p.D430E |
| BICR18_UPPER_AERODIGESTIVE_TRACT | 85701293 | 85701421 | 85701411 | 85701411 | Missense_Mutation | A | C | p.D430E |
| DAUDI_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85701293 | 85701442 | 85701425 | 85701425 | Missense_Mutation | C | G | p.V426L |
| DAUDI_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85701293 | 85701421 | 85701425 | 85701425 | Missense_Mutation | C | G | p.V426L |
| BICR18_UPPER_AERODIGESTIVE_TRACT | 85701293 | 85701442 | 85701425 | 85701425 | Missense_Mutation | C | G | p.V426L |
| BICR18_UPPER_AERODIGESTIVE_TRACT | 85701293 | 85701421 | 85701425 | 85701425 | Missense_Mutation | C | G | p.V426L |
| SNUC2A_LARGE_INTESTINE | 85711686 | 85711822 | 85711699 | 85711699 | Missense_Mutation | T | C | p.S381G |
| SLR24_KIDNEY | 85711686 | 85711822 | 85711712 | 85711712 | Missense_Mutation | T | C | p.I376M |
| SNU1040_LARGE_INTESTINE | 85711686 | 85711822 | 85711762 | 85711762 | Missense_Mutation | G | A | p.P360S |
| ATN1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85718585 | 85718626 | 85718593 | 85718593 | Missense_Mutation | G | A | p.L267F |
| HOP62_LUNG | 85718585 | 85718626 | 85718614 | 85718614 | Missense_Mutation | C | G | p.D260H |
| YT_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 85723324 | 85723435 | 85723375 | 85723375 | Missense_Mutation | C | T | p.D203N |
| HCC2998_LARGE_INTESTINE | 85733410 | 85733512 | 85733423 | 85733423 | Missense_Mutation | T | G | p.K147Q |
| HCT116_LARGE_INTESTINE | 85733410 | 85733512 | 85733486 | 85733486 | Missense_Mutation | G | A | p.R126W |
| SW948_LARGE_INTESTINE | 85737334 | 85737409 | 85737367 | 85737367 | Missense_Mutation | A | C | p.L106V |
| HEC251_ENDOMETRIUM | 85742511 | 85742653 | 85742554 | 85742554 | Missense_Mutation | T | G | p.K77T |
| HCC1438_LUNG | 85742511 | 85742653 | 85742568 | 85742568 | Missense_Mutation | C | A | p.W72C |
| KMRC1_KIDNEY | 85742511 | 85742653 | 85742635 | 85742635 | Missense_Mutation | T | C | p.N50S |
| NCIH2023_LUNG | 85779693 | 85779775 | 85779770 | 85779770 | Missense_Mutation | G | T | p.T18N |












 Gene summary
Gene summary Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez  Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 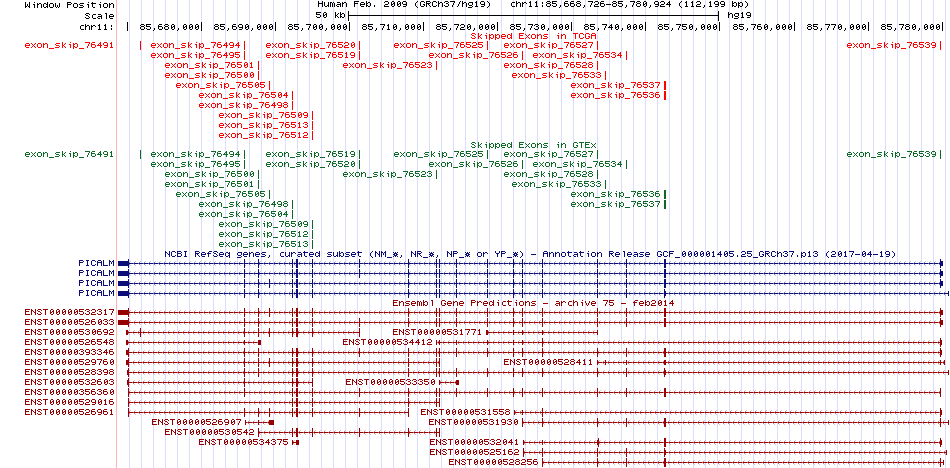
 Expression levels of gene isoforms across TCGA.
Expression levels of gene isoforms across TCGA. Expression levels of gene isoforms across GTEx.
Expression levels of gene isoforms across GTEx. Information of exkip skipping event in TCGA.
Information of exkip skipping event in TCGA. PSI values of skipped exons in TCGA.
PSI values of skipped exons in TCGA. Information of exkip skipping event in GTEx
Information of exkip skipping event in GTEx PSI values of skipped exons in GTEx.
PSI values of skipped exons in GTEx. Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms.
Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms.
Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms. Exon skipping at the protein sequence level and followed lost functional features.
Exon skipping at the protein sequence level and followed lost functional features.
 Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases.
Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases.
Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases. Lost protein functional features of individual exon skipping events in TCGA.
Lost protein functional features of individual exon skipping events in TCGA. Lost protein functional features of individual exon skipping events in GTEx.
Lost protein functional features of individual exon skipping events in GTEx. - Lollipop plot for presenting exon skipping associated SNVs.
- Lollipop plot for presenting exon skipping associated SNVs. - Differential PSIs between mutated versus non-mutated samples.
- Differential PSIs between mutated versus non-mutated samples.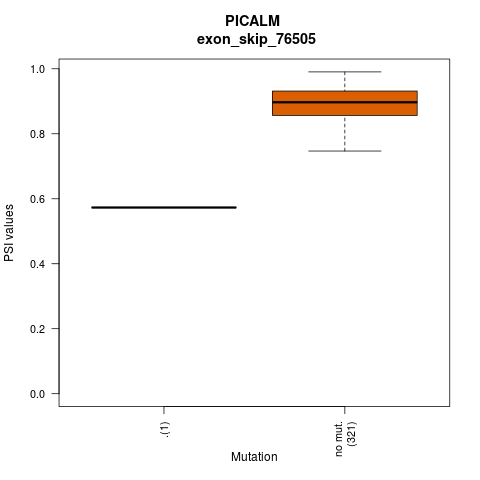
 - Non-synonymous mutations located in the skipped exons in TCGA.
- Non-synonymous mutations located in the skipped exons in TCGA. - Depth of coverage in the three exons composing exon skipping event
- Depth of coverage in the three exons composing exon skipping event - Non-synonymous mutations located in the skipped exons in CCLE.
- Non-synonymous mutations located in the skipped exons in CCLE. sQTL information located at the skipped exons.
sQTL information located at the skipped exons. Approved drugs targeting this gene.
Approved drugs targeting this gene.  Diseases associated with this gene.
Diseases associated with this gene. 









