
|
||||||
|
 | |
 | |
 | |
 | |
 | Open reading frame (ORF) annotation in the exon skipping event |
 | |
 | |
 | Splicing Quantitative Trait Loci (sQTLs) in the skipped exons |
 | Splicing Quantitative Trait Methylation (sQTM) in the skipped exon |
 | |
 |
Gene summary for SIRT1 |
 Gene summary Gene summary |
| Gene information | Gene symbol | SIRT1 | Gene ID | 23411 |
| Gene name | sirtuin 1 | |
| Synonyms | SIR2|SIR2L1|SIR2alpha | |
| Cytomap | 10q21.3 | |
| Type of gene | protein-coding | |
| Description | NAD-dependent protein deacetylase sirtuin-1SIR2-like protein 1regulatory protein SIR2 homolog 1sirtuin type 1 | |
| Modification date | 20180527 | |
| UniProtAcc | Q96EB6 | |
| Context | PubMed: SIRT1 [Title/Abstract] AND exon [Title/Abstract] AND skip [Title/Abstract] - Title (PMID) |
 Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Gene | GO ID | GO term | PubMed ID |
| SIRT1 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 12535671|15692560|20955178 |
| SIRT1 | GO:0000183 | chromatin silencing at rDNA | 18485871 |
| SIRT1 | GO:0001525 | angiogenesis | 20620956 |
| SIRT1 | GO:0002821 | positive regulation of adaptive immune response | 21890893 |
| SIRT1 | GO:0006343 | establishment of chromatin silencing | 15469825 |
| SIRT1 | GO:0006476 | protein deacetylation | 18203716|18662546|20027304|20955178 |
| SIRT1 | GO:0006974 | cellular response to DNA damage stimulus | 18203716 |
| SIRT1 | GO:0006979 | response to oxidative stress | 14976264 |
| SIRT1 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 23960241 |
| SIRT1 | GO:0007346 | regulation of mitotic cell cycle | 15692560 |
| SIRT1 | GO:0016239 | positive regulation of macroautophagy | 18296641 |
| SIRT1 | GO:0016567 | protein ubiquitination | 21841822 |
| SIRT1 | GO:0016575 | histone deacetylation | 12006491|15469825|16079181|17172643 |
| SIRT1 | GO:0031648 | protein destabilization | 20955178 |
| SIRT1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 15152190 |
| SIRT1 | GO:0034983 | peptidyl-lysine deacetylation | 15469825 |
| SIRT1 | GO:0042542 | response to hydrogen peroxide | 19934257 |
| SIRT1 | GO:0043065 | positive regulation of apoptotic process | 15152190 |
| SIRT1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 17680780 |
| SIRT1 | GO:0043433 | negative regulation of DNA binding transcription factor activity | 11672523|20955178 |
| SIRT1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 11672523 |
| SIRT1 | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 23960241 |
| SIRT1 | GO:0045348 | positive regulation of MHC class II biosynthetic process | 21890893 |
| SIRT1 | GO:0045766 | positive regulation of angiogenesis | 23960241|25217442 |
| SIRT1 | GO:0045892 | negative regulation of transcription, DNA-templated | 11672523|20074560 |
| SIRT1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 12837246|21807113 |
| SIRT1 | GO:0046628 | positive regulation of insulin receptor signaling pathway | 21241768 |
| SIRT1 | GO:0051097 | negative regulation of helicase activity | 18203716 |
| SIRT1 | GO:0070301 | cellular response to hydrogen peroxide | 20027304 |
| SIRT1 | GO:0070932 | histone H3 deacetylation | 20027304 |
| SIRT1 | GO:0071356 | cellular response to tumor necrosis factor | 15152190 |
| SIRT1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 20203304 |
| SIRT1 | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 20100829 |
| SIRT1 | GO:2000773 | negative regulation of cellular senescence | 20203304 |
| SIRT1 | GO:2000774 | positive regulation of cellular senescence | 18687677 |
Top |
Exon skipping events across known transcript of Ensembl for SIRT1 from UCSC genome browser |
 Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. * Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
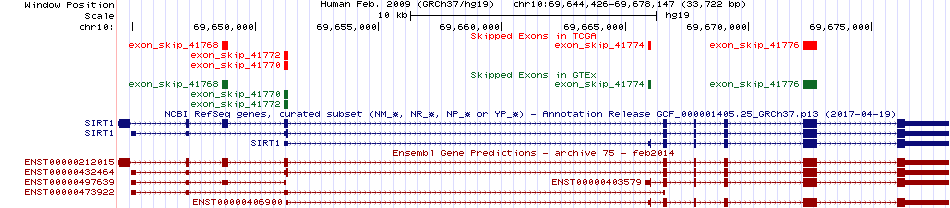 |
Top |
Gene isoform structures and expression levels for SIRT1 |
 Expression levels of gene isoforms across TCGA. Expression levels of gene isoforms across TCGA. |
 Expression levels of gene isoforms across GTEx. Expression levels of gene isoforms across GTEx. |
Top |
Exon skipping events with PSIs in TCGA for SIRT1 |
 Information of exkip skipping event in TCGA. Information of exkip skipping event in TCGA. |
| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_41768 | 10 | 69647174:69647291:69648639:69648881:69651159:69651254 | 69648639:69648881 | ENSG00000096717.7 | ENST00000212015.6,ENST00000497639.1 |
| exon_skip_41770 | 10 | 69647174:69647291:69651159:69651312:69666546:69666636 | 69651159:69651312 | ENSG00000096717.7 | ENST00000432464.1,ENST00000473922.1 |
| exon_skip_41772 | 10 | 69648639:69648881:69651159:69651312:69666546:69666636 | 69651159:69651312 | ENSG00000096717.7 | ENST00000212015.6 |
| exon_skip_41774 | 10 | 69651241:69651312:69665919:69666044:69666546:69666636 | 69665919:69666044 | ENSG00000096717.7 | ENST00000406900.1 |
| exon_skip_41776 | 10 | 69669012:69669199:69672230:69672788:69676021:69678143 | 69672230:69672788 | ENSG00000096717.7 | ENST00000432464.1,ENST00000212015.6,ENST00000406900.1,ENST00000403579.1 |
 PSI values of skipped exons in TCGA. PSI values of skipped exons in TCGA. |
Top |
Exon skipping events with PSIs in GTEx for SIRT1 |
 Information of exkip skipping event in GTEx Information of exkip skipping event in GTEx |
| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_41768 | 10 | 69647174:69647291:69648639:69648881:69651159:69651254 | 69648639:69648881 | ENSG00000096717.7 | ENST00000212015.6,ENST00000497639.1 |
| exon_skip_41770 | 10 | 69647174:69647291:69651159:69651312:69666546:69666636 | 69651159:69651312 | ENSG00000096717.7 | ENST00000432464.1,ENST00000473922.1 |
| exon_skip_41772 | 10 | 69648639:69648881:69651159:69651312:69666546:69666636 | 69651159:69651312 | ENSG00000096717.7 | ENST00000212015.6 |
| exon_skip_41774 | 10 | 69651241:69651312:69665919:69666044:69666546:69666636 | 69665919:69666044 | ENSG00000096717.7 | ENST00000406900.1 |
| exon_skip_41776 | 10 | 69669012:69669199:69672230:69672788:69676021:69678143 | 69672230:69672788 | ENSG00000096717.7 | ENST00000212015.6,ENST00000432464.1,ENST00000406900.1,ENST00000403579.1 |
 PSI values of skipped exons in GTEx. PSI values of skipped exons in GTEx. |
| * Skipped exon sequences. |
Top |
Open reading frame (ORF) annotation in the exon skipping event for SIRT1 |
 Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms. |
| ENST | Start of skipped exon | End of skipped exon | ORF |
| ENST00000212015 | 69648639 | 69648881 | Frame-shift |
| ENST00000212015 | 69651159 | 69651312 | In-frame |
| ENST00000212015 | 69672230 | 69672788 | In-frame |
 Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms. |
| ENST | Start of skipped exon | End of skipped exon | ORF |
| ENST00000212015 | 69648639 | 69648881 | Frame-shift |
| ENST00000212015 | 69651159 | 69651312 | In-frame |
| ENST00000212015 | 69672230 | 69672788 | In-frame |
Top |
Infer the effects of exon skipping event on protein functional features for SIRT1 |
 Exon skipping at the protein sequence level and followed lost functional features. Exon skipping at the protein sequence level and followed lost functional features.* Click on the image to enlarge it in a new window. |
 |
 Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases. |
| ENST | Length of mRNA | Length of AA seq. | Genomic start | Genomic end | mRNA start | mRNA end | AA start | AA end |
| ENST00000212015 | 4111 | 747 | 69651159 | 69651312 | 843 | 995 | 263 | 314 |
| ENST00000212015 | 4111 | 747 | 69672230 | 69672788 | 1411 | 1968 | 452 | 638 |
 Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases. |
| ENST | Length of mRNA | Length of AA seq. | Genomic start | Genomic end | mRNA start | mRNA end | AA start | AA end |
| ENST00000212015 | 4111 | 747 | 69651159 | 69651312 | 843 | 995 | 263 | 314 |
| ENST00000212015 | 4111 | 747 | 69672230 | 69672788 | 1411 | 1968 | 452 | 638 |
 Lost protein functional features of individual exon skipping events in TCGA. Lost protein functional features of individual exon skipping events in TCGA. |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q96EB6 | 263 | 314 | 273 | 275 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 290 | 292 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 2 | 747 | Chain | ID=PRO_0000110256;Note=NAD-dependent protein deacetylase sirtuin-1 |
| Q96EB6 | 263 | 314 | 2 | 533 | Chain | ID=PRO_0000415289;Note=SirtT1 75 kDa fragment |
| Q96EB6 | 263 | 314 | 244 | 498 | Domain | Note=Deacetylase sirtuin-type;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00236 |
| Q96EB6 | 263 | 314 | 262 | 268 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 279 | 286 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 293 | 297 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 299 | 304 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 307 | 312 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 313 | 316 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 261 | 280 | Nucleotide binding | Note=NAD;Ontology_term=ECO:0000250;evidence=ECO:0000250 |
| Q96EB6 | 263 | 314 | 2 | 268 | Region | Note=Interaction with HIST1H1E |
| Q96EB6 | 263 | 314 | 143 | 541 | Region | Note=Interaction with CCAR2 |
| Q96EB6 | 263 | 314 | 276 | 278 | Turn | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4IG9 |
| Q96EB6 | 452 | 638 | 454 | 639 | Alternative sequence | ID=VSP_042189;Note=In isoform 2. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q96EB6 | 452 | 638 | 455 | 460 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 461 | 467 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 475 | 480 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 515 | 517 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 526 | 531 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 554 | 560 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 582 | 589 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 599 | 604 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 629 | 633 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 482 | 482 | Binding site | Note=NAD%3B via amide nitrogen;Ontology_term=ECO:0000250;evidence=ECO:0000250 |
| Q96EB6 | 452 | 638 | 2 | 747 | Chain | ID=PRO_0000110256;Note=NAD-dependent protein deacetylase sirtuin-1 |
| Q96EB6 | 452 | 638 | 2 | 533 | Chain | ID=PRO_0000415289;Note=SirtT1 75 kDa fragment |
| Q96EB6 | 452 | 638 | 244 | 498 | Domain | Note=Deacetylase sirtuin-type;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00236 |
| Q96EB6 | 452 | 638 | 451 | 454 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 482 | 493 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 495 | 500 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 502 | 507 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 542 | 548 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 571 | 573 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 574 | 577 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 606 | 616 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 618 | 621 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 530 | 530 | Modified residue | Note=Phosphothreonine%3B by DYRK1A%2C DYRK3 and MAPK8;Ontology_term=ECO:0000244,ECO:0000269,ECO:0000269;evidence=ECO:0000244|PubMed:19690332,ECO:0000269|PubMed:19107194,ECO:0000269|PubMed:20027304;Dbxref=PMID:19690332,PMID:19107194,PMID:20027304 |
| Q96EB6 | 452 | 638 | 535 | 535 | Modified residue | Note=Phosphoserine;Ontology_term=ECO:0000244;evidence=ECO:0000244|PubMed:19690332;Dbxref=PMID:19690332 |
| Q96EB6 | 452 | 638 | 544 | 544 | Modified residue | Note=Phosphothreonine;Ontology_term=ECO:0000305;evidence=ECO:0000305|PubMed:19107194;Dbxref=PMID:19107194 |
| Q96EB6 | 452 | 638 | 545 | 545 | Modified residue | Note=Phosphoserine;Ontology_term=ECO:0000305;evidence=ECO:0000305|PubMed:19107194;Dbxref=PMID:19107194 |
| Q96EB6 | 452 | 638 | 474 | 474 | Mutagenesis | Note=Abolishes phosphorylation at Ser-47%2C restores deacetylation activity and inhibits DNA damage-induced apoptosis. F->A;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:21471201;Dbxref=PMID:21471201 |
| Q96EB6 | 452 | 638 | 530 | 530 | Mutagenesis | Note=Greatly diminishes phosphorylation by MAPK8%3B when associated with A-27 and A-47. T->A;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19107194,ECO:0000269|PubMed:20027304;Dbxref=PMID:19107194,PMID:20027304 |
| Q96EB6 | 452 | 638 | 530 | 530 | Mutagenesis | Note=Reduces in vitro phosphorylation by CDK1. Impairs cell proliferation and cell cycle progression%3B when associated with A-540. T->A;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19107194,ECO:0000269|PubMed:20027304;Dbxref=PMID:191 |
| Q96EB6 | 452 | 638 | 540 | 540 | Mutagenesis | Note=Reduces in vitro phosphorylation by CDK1. Impairs cell proliferation and cell cycle progression%3B when associated with A-530. S->A;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:19107194;Dbxref=PMID:19107194 |
| Q96EB6 | 452 | 638 | 484 | 484 | Natural variant | ID=VAR_051976;Note=V->D;Dbxref=dbSNP:rs1063111 |
| Q96EB6 | 452 | 638 | 465 | 467 | Nucleotide binding | Note=NAD;Ontology_term=ECO:0000250;evidence=ECO:0000250 |
| Q96EB6 | 452 | 638 | 143 | 541 | Region | Note=Interaction with CCAR2 |
| Q96EB6 | 452 | 638 | 538 | 540 | Region | Note=Phosphorylated at one of three serine residues |
| Q96EB6 | 452 | 638 | 549 | 551 | Turn | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
 Lost protein functional features of individual exon skipping events in GTEx. Lost protein functional features of individual exon skipping events in GTEx. |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q96EB6 | 263 | 314 | 273 | 275 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 290 | 292 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 2 | 747 | Chain | ID=PRO_0000110256;Note=NAD-dependent protein deacetylase sirtuin-1 |
| Q96EB6 | 263 | 314 | 2 | 533 | Chain | ID=PRO_0000415289;Note=SirtT1 75 kDa fragment |
| Q96EB6 | 263 | 314 | 244 | 498 | Domain | Note=Deacetylase sirtuin-type;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00236 |
| Q96EB6 | 263 | 314 | 262 | 268 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 279 | 286 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 293 | 297 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 299 | 304 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 307 | 312 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 313 | 316 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 263 | 314 | 261 | 280 | Nucleotide binding | Note=NAD;Ontology_term=ECO:0000250;evidence=ECO:0000250 |
| Q96EB6 | 263 | 314 | 2 | 268 | Region | Note=Interaction with HIST1H1E |
| Q96EB6 | 263 | 314 | 143 | 541 | Region | Note=Interaction with CCAR2 |
| Q96EB6 | 263 | 314 | 276 | 278 | Turn | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4IG9 |
| Q96EB6 | 452 | 638 | 454 | 639 | Alternative sequence | ID=VSP_042189;Note=In isoform 2. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q96EB6 | 452 | 638 | 455 | 460 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 461 | 467 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 475 | 480 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 515 | 517 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 526 | 531 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 554 | 560 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 582 | 589 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 599 | 604 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 629 | 633 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 482 | 482 | Binding site | Note=NAD%3B via amide nitrogen;Ontology_term=ECO:0000250;evidence=ECO:0000250 |
| Q96EB6 | 452 | 638 | 2 | 747 | Chain | ID=PRO_0000110256;Note=NAD-dependent protein deacetylase sirtuin-1 |
| Q96EB6 | 452 | 638 | 2 | 533 | Chain | ID=PRO_0000415289;Note=SirtT1 75 kDa fragment |
| Q96EB6 | 452 | 638 | 244 | 498 | Domain | Note=Deacetylase sirtuin-type;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00236 |
| Q96EB6 | 452 | 638 | 451 | 454 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 482 | 493 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 495 | 500 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4KXQ |
| Q96EB6 | 452 | 638 | 502 | 507 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 542 | 548 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 571 | 573 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 574 | 577 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 606 | 616 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 618 | 621 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
| Q96EB6 | 452 | 638 | 530 | 530 | Modified residue | Note=Phosphothreonine%3B by DYRK1A%2C DYRK3 and MAPK8;Ontology_term=ECO:0000244,ECO:0000269,ECO:0000269;evidence=ECO:0000244|PubMed:19690332,ECO:0000269|PubMed:19107194,ECO:0000269|PubMed:20027304;Dbxref=PMID:19690332,PMID:19107194,PMID:20027304 |
| Q96EB6 | 452 | 638 | 535 | 535 | Modified residue | Note=Phosphoserine;Ontology_term=ECO:0000244;evidence=ECO:0000244|PubMed:19690332;Dbxref=PMID:19690332 |
| Q96EB6 | 452 | 638 | 544 | 544 | Modified residue | Note=Phosphothreonine;Ontology_term=ECO:0000305;evidence=ECO:0000305|PubMed:19107194;Dbxref=PMID:19107194 |
| Q96EB6 | 452 | 638 | 545 | 545 | Modified residue | Note=Phosphoserine;Ontology_term=ECO:0000305;evidence=ECO:0000305|PubMed:19107194;Dbxref=PMID:19107194 |
| Q96EB6 | 452 | 638 | 474 | 474 | Mutagenesis | Note=Abolishes phosphorylation at Ser-47%2C restores deacetylation activity and inhibits DNA damage-induced apoptosis. F->A;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:21471201;Dbxref=PMID:21471201 |
| Q96EB6 | 452 | 638 | 530 | 530 | Mutagenesis | Note=Greatly diminishes phosphorylation by MAPK8%3B when associated with A-27 and A-47. T->A;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19107194,ECO:0000269|PubMed:20027304;Dbxref=PMID:19107194,PMID:20027304 |
| Q96EB6 | 452 | 638 | 530 | 530 | Mutagenesis | Note=Reduces in vitro phosphorylation by CDK1. Impairs cell proliferation and cell cycle progression%3B when associated with A-540. T->A;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19107194,ECO:0000269|PubMed:20027304;Dbxref=PMID:191 |
| Q96EB6 | 452 | 638 | 540 | 540 | Mutagenesis | Note=Reduces in vitro phosphorylation by CDK1. Impairs cell proliferation and cell cycle progression%3B when associated with A-530. S->A;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:19107194;Dbxref=PMID:19107194 |
| Q96EB6 | 452 | 638 | 484 | 484 | Natural variant | ID=VAR_051976;Note=V->D;Dbxref=dbSNP:rs1063111 |
| Q96EB6 | 452 | 638 | 465 | 467 | Nucleotide binding | Note=NAD;Ontology_term=ECO:0000250;evidence=ECO:0000250 |
| Q96EB6 | 452 | 638 | 143 | 541 | Region | Note=Interaction with CCAR2 |
| Q96EB6 | 452 | 638 | 538 | 540 | Region | Note=Phosphorylated at one of three serine residues |
| Q96EB6 | 452 | 638 | 549 | 551 | Turn | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5BTR |
Top |
SNVs in the skipped exons for SIRT1 |
 - Lollipop plot for presenting exon skipping associated SNVs. - Lollipop plot for presenting exon skipping associated SNVs.* Click on the image to enlarge it in a new window. |
 - Differential PSIs between mutated versus non-mutated samples. - Differential PSIs between mutated versus non-mutated samples. |
 - Non-synonymous mutations located in the skipped exons in TCGA. - Non-synonymous mutations located in the skipped exons in TCGA. |
| Cancer type | Sample | ESID | Skipped exon start | Skipped exon end | Mutation start | Mutation end | Mutation type | Reference seq | Mutation seq | AAchange |
| KICH | TCGA-KO-8410-01 | exon_skip_41768 | 69648640 | 69648881 | 69648852 | 69648852 | Frame_Shift_Del | A | - | p.K254fs |
| PRAD | TCGA-KK-A59V-01 | exon_skip_41768 | 69648640 | 69648881 | 69648852 | 69648852 | Frame_Shift_Del | A | - | p.C253X |
| KIRC | TCGA-CZ-5982-01 | exon_skip_41776 | 69672231 | 69672788 | 69672600 | 69672600 | Frame_Shift_Del | A | - | p.E576fs |
| KIRC | TCGA-CZ-5982-01 | exon_skip_41776 | 69672231 | 69672788 | 69672600 | 69672600 | Frame_Shift_Del | A | - | p.E577fs |
| LIHC | TCGA-BC-A112-01 | exon_skip_41776 | 69672231 | 69672788 | 69672559 | 69672560 | Frame_Shift_Ins | - | A | p.*563fs |
| SARC | TCGA-QC-A7B5-01 | exon_skip_41768 | 69648640 | 69648881 | 69648754 | 69648754 | Nonsense_Mutation | G | A | p.W221* |
| CESC | TCGA-IR-A3LH-01 | exon_skip_41768 | 69648640 | 69648881 | 69648778 | 69648778 | Nonsense_Mutation | C | G | p.S229* |
| UCEC | TCGA-B5-A0JY-01 | exon_skip_41776 | 69672231 | 69672788 | 69672272 | 69672272 | Nonsense_Mutation | G | T | p.E172* |
| STAD | TCGA-FP-8099-01 | exon_skip_41776 | 69672231 | 69672788 | 69672419 | 69672419 | Nonsense_Mutation | C | T | p.R516* |
| STAD | TCGA-FP-8099-01 | exon_skip_41776 | 69672231 | 69672788 | 69672419 | 69672419 | Nonsense_Mutation | C | T | p.R516X |
| UCEC | TCGA-AX-A05Z-01 | exon_skip_41776 | 69672231 | 69672788 | 69672431 | 69672431 | Nonsense_Mutation | G | T | p.E225* |
 - Depth of coverage in the three exons composing exon skipping event - Depth of coverage in the three exons composing exon skipping event |
| Depth of coverage in three exons | Mutation description |
 - Non-synonymous mutations located in the skipped exons in CCLE. - Non-synonymous mutations located in the skipped exons in CCLE. |
| Sample | Skipped exon start | Skipped exon end | Mutation start | Mutation end | Mutation type | Reference seq | Mutation seq | AAchange |
| IM95_STOMACH | 69648640 | 69648881 | 69648795 | 69648795 | Frame_Shift_Del | A | - | p.K236fs |
| DV90_LUNG | 69648640 | 69648881 | 69648852 | 69648852 | Frame_Shift_Del | A | - | p.K255fs |
| SNU1033_LARGE_INTESTINE | 69648640 | 69648881 | 69648794 | 69648795 | Frame_Shift_Ins | - | A | p.K235fs |
| SNGM_ENDOMETRIUM | 69648640 | 69648881 | 69648688 | 69648688 | Missense_Mutation | G | A | p.R199Q |
| C75_LARGE_INTESTINE | 69648640 | 69648881 | 69648712 | 69648712 | Missense_Mutation | C | A | p.P207Q |
| SNU81_LARGE_INTESTINE | 69648640 | 69648881 | 69648805 | 69648805 | Missense_Mutation | A | C | p.K238T |
| SCC4_UPPER_AERODIGESTIVE_TRACT | 69651160 | 69651312 | 69651179 | 69651179 | Missense_Mutation | T | A | p.I270K |
| CTV1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 69672231 | 69672788 | 69672305 | 69672305 | Missense_Mutation | C | G | p.L478V |
| TF1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 69672231 | 69672788 | 69672312 | 69672312 | Missense_Mutation | G | A | p.G480E |
| HEC251_ENDOMETRIUM | 69672231 | 69672788 | 69672339 | 69672339 | Missense_Mutation | T | C | p.L489S |
| NCIH1836_LUNG | 69672231 | 69672788 | 69672357 | 69672357 | Missense_Mutation | G | A | p.G495D |
| SNUC2A_LARGE_INTESTINE | 69672231 | 69672788 | 69672366 | 69672366 | Missense_Mutation | C | T | p.A498V |
| JHUEM7_ENDOMETRIUM | 69672231 | 69672788 | 69672420 | 69672420 | Missense_Mutation | G | A | p.R516Q |
| HPBALL_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 69672231 | 69672788 | 69672747 | 69672747 | Missense_Mutation | C | T | p.P625L |
| NCIH146_LUNG | 69672231 | 69672788 | 69672769 | 69672769 | Missense_Mutation | G | T | p.Q632H |
| HCC2998_LARGE_INTESTINE | 69672231 | 69672788 | 69672431 | 69672431 | Nonsense_Mutation | G | T | p.E520* |
Top |
Splicing Quantitative Trait Loci (sQTL) in the exon skipping event for SIRT1 |
 sQTL information located at the skipped exons. sQTL information located at the skipped exons. |
| Exon skip ID | Chromosome | Three exons | Skippped exon | ENST | Cancer type | SNP id | Location | DNA change (ref/var) | P-value |
Top |
Splicing Quantitative Trait Methylation (sQTM) in the skipped exon for SIRT1 |
Top |
Survival analysis of Splicing Quantitative Trait Methylation (sQTM) in the skipped exon for SIRT1 |
Top |
RelatedDrugs for SIRT1 |
 Approved drugs targeting this gene. Approved drugs targeting this gene. (DrugBank Version 5.1.0 2018-04-02) |
| Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
RelatedDiseases for SIRT1 |
 Diseases associated with this gene. Diseases associated with this gene. (DisGeNet 4.0) |
| Gene | Disease ID | Disease name | # pubmeds | Source |
| SIRT1 | C0015695 | Fatty Liver | 5 | CTD_human |
| SIRT1 | C0021655 | Insulin Resistance | 5 | CTD_human |
| SIRT1 | C0009319 | Colitis | 3 | CTD_human |
| SIRT1 | C0005586 | Bipolar Disorder | 2 | PSYGENET |
| SIRT1 | C0011853 | Diabetes Mellitus, Experimental | 2 | CTD_human |
| SIRT1 | C0036341 | Schizophrenia | 2 | PSYGENET |
| SIRT1 | C0524851 | Neurodegenerative Disorders | 2 | CTD_human |
| SIRT1 | C0525045 | Mood Disorders | 2 | PSYGENET |
| SIRT1 | C0004153 | Atherosclerosis | 1 | CTD_human |
| SIRT1 | C0004364 | Autoimmune Diseases | 1 | CTD_human |
| SIRT1 | C0006118 | Brain Neoplasms | 1 | CTD_human |
| SIRT1 | C0011303 | Demyelinating Diseases | 1 | CTD_human |
| SIRT1 | C0011849 | Diabetes Mellitus | 1 | CTD_human |
| SIRT1 | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| SIRT1 | C0011884 | Diabetic Retinopathy | 1 | CTD_human |
| SIRT1 | C0014072 | Experimental Autoimmune Encephalomyelitis | 1 | CTD_human |
| SIRT1 | C0016059 | Fibrosis | 1 | CTD_human |
| SIRT1 | C0018799 | Heart Diseases | 1 | CTD_human |
| SIRT1 | C0018801 | Heart failure | 1 | CTD_human |
| SIRT1 | C0019693 | HIV Infections | 1 | CTD_human |
| SIRT1 | C0020564 | Hypertrophy | 1 | CTD_human |
| SIRT1 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| SIRT1 | C0027055 | Myocardial Reperfusion Injury | 1 | CTD_human |
| SIRT1 | C0027746 | Nerve Degeneration | 1 | CTD_human |
| SIRT1 | C0028754 | Obesity | 1 | CTD_human |
| SIRT1 | C0032285 | Pneumonia | 1 | CTD_human |
| SIRT1 | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| SIRT1 | C0033937 | Psychoses, Drug | 1 | PSYGENET |
| SIRT1 | C0035309 | Retinal Diseases | 1 | CTD_human |
| SIRT1 | C0036421 | Systemic Scleroderma | 1 | CTD_human |
| SIRT1 | C0036939 | Shared Paranoid Disorder | 1 | PSYGENET |
| SIRT1 | C0040053 | Thrombosis | 1 | CTD_human |
| SIRT1 | C0043020 | Wallerian Degeneration | 1 | CTD_human |
| SIRT1 | C0085762 | Alcohol abuse | 1 | PSYGENET |
| SIRT1 | C0155862 | Streptococcal pneumonia | 1 | CTD_human |
| SIRT1 | C0271650 | Impaired glucose tolerance | 1 | CTD_human |
| SIRT1 | C0273115 | Lung Injury | 1 | CTD_human |
| SIRT1 | C0400966 | Non-alcoholic Fatty Liver Disease | 1 | CTD_human |
| SIRT1 | C0522224 | Paralysed | 1 | CTD_human |
| SIRT1 | C0524620 | Metabolic Syndrome X | 1 | CTD_human |
| SIRT1 | C0751955 | Brain Infarction | 1 | CTD_human |
| SIRT1 | C0919532 | Genomic Instability | 1 | CTD_human |
| SIRT1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| SIRT1 | C2609414 | Acute kidney injury | 1 | CTD_human |
| SIRT1 | C2931673 | Ceroid lipofuscinosis, neuronal 1, infantile | 1 | CTD_human |
