| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_81230 | 12 | 31226842:31227025:31231323:31231471:31236746:31236995 | 31231323:31231471 | ENSG00000013573.12 | ENST00000536265.1,ENST00000407793.2,ENST00000251758.5,ENST00000535158.1,ENST00000350437.4,ENST00000435753.2,ENST00000542129.1,ENST00000438391.2,ENST00000539049.1 |
| exon_skip_81231 | 12 | 31226842:31227025:31231396:31231471:31236746:31236995 | 31231396:31231471 | ENSG00000013573.12 | ENST00000415475.2 |
| exon_skip_81232 | 12 | 31227197:31227261:31230878:31231222:31231323:31231471 | 31230878:31231222 | ENSG00000013573.12 | ENST00000535317.1 |
| exon_skip_81234 | 12 | 31227197:31227261:31231323:31231471:31236746:31236995 | 31231323:31231471 | ENSG00000013573.12 | ENST00000542244.1 |
| exon_skip_81237 | 12 | 31236900:31236995:31237516:31237603:31237902:31238002 | 31237516:31237603 | ENSG00000013573.12 | ENST00000536265.1,ENST00000407793.2,ENST00000251758.5,ENST00000535158.1,ENST00000350437.4,ENST00000542838.1,ENST00000435753.2,ENST00000415475.2,ENST00000542129.1,ENST00000540935.1,ENST00000228264.6,ENST00000543756.1,ENST00000539049.1,ENST00000545668.1 |
| exon_skip_81240 | 12 | 31236900:31236995:31237902:31238060:31240871:31240917 | 31237902:31238060 | ENSG00000013573.12 | ENST00000438391.2 |
| exon_skip_81241 | 12 | 31237516:31237603:31237731:31237845:31237943:31238060 | 31237731:31237845 | ENSG00000013573.12 | ENST00000542244.1 |
| exon_skip_81243 | 12 | 31237516:31237603:31237902:31238060:31240871:31240917 | 31237902:31238060 | ENSG00000013573.12 | ENST00000536265.1,ENST00000407793.2,ENST00000535158.1,ENST00000350437.4,ENST00000542838.1,ENST00000415475.2,ENST00000542129.1,ENST00000540935.1,ENST00000228264.6,ENST00000539049.1,ENST00000545668.1 |
| exon_skip_81248 | 12 | 31237943:31238060:31238958:31239146:31240871:31240917 | 31238958:31239146 | ENSG00000013573.12 | ENST00000251758.5,ENST00000435753.2 |
| exon_skip_81249 | 12 | 31237943:31238060:31240871:31240917:31241977:31242045 | 31240871:31240917 | ENSG00000013573.12 | ENST00000536265.1,ENST00000407793.2,ENST00000535158.1,ENST00000350437.4,ENST00000542838.1,ENST00000415475.2,ENST00000542129.1,ENST00000542244.1,ENST00000540935.1,ENST00000228264.6,ENST00000438391.2,ENST00000539049.1,ENST00000545668.1 |
| exon_skip_81250 | 12 | 31242002:31242085:31242336:31242424:31242819:31242933 | 31242336:31242424 | ENSG00000013573.12 | ENST00000407793.2,ENST00000251758.5,ENST00000350437.4,ENST00000542838.1,ENST00000228264.6,ENST00000438391.2,ENST00000545668.1,ENST00000545717.1 |
| exon_skip_81254 | 12 | 31245795:31245826:31246178:31246258:31247523:31247567 | 31246178:31246258 | ENSG00000013573.12 | ENST00000536265.1,ENST00000407793.2,ENST00000251758.5,ENST00000350437.4,ENST00000542838.1,ENST00000536580.1,ENST00000435753.2,ENST00000542129.1,ENST00000228264.6,ENST00000539049.1,ENST00000539673.1,ENST00000545668.1 |
| exon_skip_81257 | 12 | 31249847:31249924:31250818:31250931:31253568:31253641 | 31250818:31250931 | ENSG00000013573.12 | ENST00000536265.1,ENST00000407793.2,ENST00000251758.5,ENST00000350437.4,ENST00000542838.1,ENST00000435753.2,ENST00000228264.6,ENST00000539699.1,ENST00000545668.1 |
| exon_skip_81264 | 12 | 31250818:31250931:31255176:31255245:31255360:31255461 | 31255176:31255245 | ENSG00000013573.12 | ENST00000542661.1 |
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENSG00000013573.12 | ENST00000350437.4 |
| exon_skip_81272 | 12 | 31254006:31254064:31254766:31254916:31255176:31255195 | 31254766:31254916 | ENSG00000013573.12 | ENST00000536265.1,ENST00000407793.2,ENST00000251758.5,ENST00000542838.1,ENST00000435753.2,ENST00000228264.6,ENST00000545668.1 |
| exon_skip_81273 | 12 | 31255384:31255461:31255869:31255954:31256250:31256329 | 31255869:31255954 | ENSG00000013573.12 | ENST00000536265.1,ENST00000539702.1,ENST00000251758.5,ENST00000350437.4,ENST00000542838.1,ENST00000435753.2,ENST00000228264.6 |
| exon_skip_81277 | 12 | 31255384:31255461:31255908:31255954:31256510:31256665 | 31255908:31255954 | ENSG00000013573.12 | ENST00000545115.1 |
| exon_skip_81279 | 12 | 31255908:31255954:31256250:31256329:31256510:31256665 | 31256250:31256329 | ENSG00000013573.12 | ENST00000536265.1,ENST00000539702.1,ENST00000251758.5,ENST00000350437.4,ENST00000542838.1,ENST00000435753.2,ENST00000228264.6 |
| exon_skip_81281 | 12 | 31255908:31255954:31256255:31256329:31256510:31256665 | 31256255:31256329 | ENSG00000013573.12 | ENST00000407793.2,ENST00000538345.1,ENST00000545668.1 |
| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_81230 | 12 | 31226842:31227025:31231323:31231471:31236746:31236995 | 31231323:31231471 | ENSG00000013573.12 | ENST00000407793.2,ENST00000251758.5,ENST00000542129.1,ENST00000438391.2,ENST00000435753.2,ENST00000539049.1,ENST00000536265.1,ENST00000350437.4,ENST00000535158.1 |
| exon_skip_81231 | 12 | 31226842:31227025:31231396:31231471:31236746:31236995 | 31231396:31231471 | ENSG00000013573.12 | ENST00000415475.2 |
| exon_skip_81232 | 12 | 31227197:31227261:31230878:31231222:31231323:31231471 | 31230878:31231222 | ENSG00000013573.12 | ENST00000535317.1 |
| exon_skip_81234 | 12 | 31227197:31227261:31231323:31231471:31236746:31236995 | 31231323:31231471 | ENSG00000013573.12 | ENST00000542244.1 |
| exon_skip_81237 | 12 | 31236900:31236995:31237516:31237603:31237902:31238002 | 31237516:31237603 | ENSG00000013573.12 | ENST00000542838.1,ENST00000407793.2,ENST00000251758.5,ENST00000542129.1,ENST00000228264.6,ENST00000435753.2,ENST00000540935.1,ENST00000539049.1,ENST00000536265.1,ENST00000415475.2,ENST00000545668.1,ENST00000350437.4,ENST00000535158.1,ENST00000543756.1 |
| exon_skip_81240 | 12 | 31236900:31236995:31237902:31238060:31240871:31240917 | 31237902:31238060 | ENSG00000013573.12 | ENST00000438391.2 |
| exon_skip_81241 | 12 | 31237516:31237603:31237731:31237845:31237943:31238060 | 31237731:31237845 | ENSG00000013573.12 | ENST00000542244.1 |
| exon_skip_81243 | 12 | 31237516:31237603:31237902:31238060:31240871:31240917 | 31237902:31238060 | ENSG00000013573.12 | ENST00000542838.1,ENST00000407793.2,ENST00000542129.1,ENST00000228264.6,ENST00000540935.1,ENST00000539049.1,ENST00000536265.1,ENST00000415475.2,ENST00000545668.1,ENST00000350437.4,ENST00000535158.1 |
| exon_skip_81248 | 12 | 31237943:31238060:31238958:31239146:31240871:31240917 | 31238958:31239146 | ENSG00000013573.12 | ENST00000251758.5,ENST00000435753.2 |
| exon_skip_81250 | 12 | 31242002:31242085:31242336:31242424:31242819:31242933 | 31242336:31242424 | ENSG00000013573.12 | ENST00000542838.1,ENST00000407793.2,ENST00000251758.5,ENST00000228264.6,ENST00000438391.2,ENST00000545668.1,ENST00000350437.4,ENST00000545717.1 |
| exon_skip_81254 | 12 | 31245795:31245826:31246178:31246258:31247523:31247567 | 31246178:31246258 | ENSG00000013573.12 | ENST00000542838.1,ENST00000407793.2,ENST00000251758.5,ENST00000542129.1,ENST00000228264.6,ENST00000435753.2,ENST00000539049.1,ENST00000536265.1,ENST00000545668.1,ENST00000350437.4,ENST00000536580.1,ENST00000539673.1 |
| exon_skip_81257 | 12 | 31249847:31249924:31250818:31250931:31253568:31253641 | 31250818:31250931 | ENSG00000013573.12 | ENST00000542838.1,ENST00000407793.2,ENST00000251758.5,ENST00000228264.6,ENST00000435753.2,ENST00000536265.1,ENST00000545668.1,ENST00000350437.4,ENST00000539699.1 |
| exon_skip_81264 | 12 | 31250818:31250931:31255176:31255245:31255360:31255461 | 31255176:31255245 | ENSG00000013573.12 | ENST00000542661.1 |
| exon_skip_81272 | 12 | 31254006:31254064:31254766:31254916:31255176:31255195 | 31254766:31254916 | ENSG00000013573.12 | ENST00000542838.1,ENST00000407793.2,ENST00000251758.5,ENST00000228264.6,ENST00000435753.2,ENST00000536265.1,ENST00000545668.1 |
| exon_skip_81273 | 12 | 31255384:31255461:31255869:31255954:31256250:31256329 | 31255869:31255954 | ENSG00000013573.12 | ENST00000542838.1,ENST00000251758.5,ENST00000228264.6,ENST00000435753.2,ENST00000536265.1,ENST00000350437.4,ENST00000539702.1 |
| exon_skip_81277 | 12 | 31255384:31255461:31255908:31255954:31256510:31256665 | 31255908:31255954 | ENSG00000013573.12 | ENST00000545115.1 |
| exon_skip_81279 | 12 | 31255908:31255954:31256250:31256329:31256510:31256665 | 31256250:31256329 | ENSG00000013573.12 | ENST00000542838.1,ENST00000251758.5,ENST00000228264.6,ENST00000435753.2,ENST00000536265.1,ENST00000350437.4,ENST00000539702.1 |
| exon_skip_81281 | 12 | 31255908:31255954:31256255:31256329:31256510:31256665 | 31256255:31256329 | ENSG00000013573.12 | ENST00000407793.2,ENST00000545668.1,ENST00000538345.1 |
| Exon skip ID | Chromosome | Three exons | Skippped exon | ENST | Cancer type | SNP id | Location | DNA change (ref/var) | P-value |
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | GBM | rs1046456 | chr12:31253995 | C/T | 3.99e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | GBM | rs1046456 | chr12:31253995 | C/T | 4.17e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | CESC | rs1046456 | chr12:31253995 | C/T | 2.68e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | CESC | rs1046456 | chr12:31253995 | C/T | 2.82e-08
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | CESC | rs1046456 | chr12:31253995 | C/T | 4.42e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | CESC | rs1046456 | chr12:31253995 | C/T | 5.30e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | COAD | rs1046456 | chr12:31253995 | C/T | 5.20e-19
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | COAD | rs1046456 | chr12:31253995 | C/T | 5.05e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | COAD | rs1046456 | chr12:31253995 | C/T | 2.35e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | COAD | rs1046456 | chr12:31253995 | C/T | 8.54e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | COAD | rs1046456 | chr12:31253995 | C/T | 9.79e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 1.66e-13
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 9.55e-08
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 3.99e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 1.29e-06
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 1.31e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 3.15e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 5.18e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BLCA | rs1046456 | chr12:31253995 | C/T | 8.89e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | ESCA | rs1046456 | chr12:31253995 | C/T | 4.13e-14
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | ESCA | rs1046456 | chr12:31253995 | C/T | 1.93e-09
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 2.89e-17
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 4.16e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 6.09e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 6.52e-08
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 1.14e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 9.63e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 9.66e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 1.88e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 2.59e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | HNSC | rs1046456 | chr12:31253995 | C/T | 1.18e-03
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 4.52e-27
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 8.75e-27
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 5.47e-24
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 1.99e-22
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 8.76e-18
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 2.07e-15
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 3.11e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 1.03e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 2.30e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 3.72e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | BRCA | rs1046456 | chr12:31253995 | C/T | 2.86e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KICH | rs1046456 | chr12:31253995 | C/T | 6.33e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KICH | rs1046456 | chr12:31253995 | C/T | 9.47e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KICH | rs1046456 | chr12:31253995 | C/T | 1.35e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KICH | rs1046456 | chr12:31253995 | C/T | 1.56e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRP | rs1046456 | chr12:31253995 | C/T | 7.56e-22
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRP | rs1046456 | chr12:31253995 | C/T | 1.39e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRP | rs1046456 | chr12:31253995 | C/T | 7.65e-11
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRP | rs1046456 | chr12:31253995 | C/T | 2.79e-06
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 1.09e-21
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 6.02e-18
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 2.31e-16
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 4.16e-14
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 2.58e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 7.05e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 6.41e-10
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 6.42e-10
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 7.36e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LGG | rs1046456 | chr12:31253995 | C/T | 8.96e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 7.24e-24
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 8.92e-13
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 7.06e-10
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 1.84e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 2.73e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 1.09e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 1.93e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 8.77e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | KIRC | rs1046456 | chr12:31253995 | C/T | 8.81e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUAD | rs1046456 | chr12:31253995 | C/T | 6.40e-24
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUAD | rs1046456 | chr12:31253995 | C/T | 2.02e-17
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUAD | rs1046456 | chr12:31253995 | C/T | 4.43e-14
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUAD | rs1046456 | chr12:31253995 | C/T | 3.36e-11
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUAD | rs1046456 | chr12:31253995 | C/T | 3.82e-11
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUAD | rs1046456 | chr12:31253995 | C/T | 1.85e-06
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUAD | rs1046456 | chr12:31253995 | C/T | 4.53e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LIHC | rs1046456 | chr12:31253995 | C/T | 1.44e-03
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUSC | rs1046456 | chr12:31253995 | C/T | 2.19e-19
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUSC | rs1046456 | chr12:31253995 | C/T | 2.18e-11
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUSC | rs1046456 | chr12:31253995 | C/T | 2.18e-11
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUSC | rs1046456 | chr12:31253995 | C/T | 2.44e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUSC | rs1046456 | chr12:31253995 | C/T | 3.03e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUSC | rs1046456 | chr12:31253995 | C/T | 3.69e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | LUSC | rs1046456 | chr12:31253995 | C/T | 9.78e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | OV | rs1046456 | chr12:31253995 | C/T | 6.49e-20
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | OV | rs1046456 | chr12:31253995 | C/T | 4.69e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | OV | rs1046456 | chr12:31253995 | C/T | 6.39e-06
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | OV | rs1046456 | chr12:31253995 | C/T | 2.35e-03
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PCPG | rs1046456 | chr12:31253995 | C/T | 3.48e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PAAD | rs1046456 | chr12:31253995 | C/T | 2.54e-08
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PAAD | rs1046456 | chr12:31253995 | C/T | 3.63e-08
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PAAD | rs1046456 | chr12:31253995 | C/T | 4.04e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PAAD | rs1046456 | chr12:31253995 | C/T | 3.26e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PAAD | rs1046456 | chr12:31253995 | C/T | 4.51e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PAAD | rs1046456 | chr12:31253995 | C/T | 9.17e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 1.01e-36
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 3.06e-26
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 1.12e-20
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 2.88e-14
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 5.01e-11
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 8.08e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 5.85e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | PRAD | rs1046456 | chr12:31253995 | C/T | 3.24e-03
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 1.06e-10
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 3.04e-09
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 2.56e-07
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 2.01e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 6.16e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 6.16e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 6.00e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SARC | rs1046456 | chr12:31253995 | C/T | 9.32e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THYM | rs1046456 | chr12:31253995 | C/T | 1.34e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THYM | rs1046456 | chr12:31253995 | C/T | 8.55e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THYM | rs1046456 | chr12:31253995 | C/T | 5.76e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | TGCT | rs1046456 | chr12:31253995 | C/T | 7.47e-15
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | TGCT | rs1046456 | chr12:31253995 | C/T | 7.68e-09
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | TGCT | rs1046456 | chr12:31253995 | C/T | 5.77e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | TGCT | rs1046456 | chr12:31253995 | C/T | 3.22e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | SKCM | rs1046456 | chr12:31253995 | C/T | 5.60e-05
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | STAD | rs1046456 | chr12:31253995 | C/T | 6.25e-12
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | STAD | rs1046456 | chr12:31253995 | C/T | 1.71e-09
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 1.96e-31
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 1.72e-27
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 5.33e-11
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 9.12e-10
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 1.24e-06
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 2.48e-06
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 1.55e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 1.56e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | THCA | rs1046456 | chr12:31253995 | C/T | 2.38e-03
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | UVM | rs1046456 | chr12:31253995 | C/T | 1.02e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | UVM | rs1046456 | chr12:31253995 | C/T | 1.84e-04
|
| exon_skip_81267 | 12 | 31253568:31253641:31253960:31254064:31255176:31255195 | 31253960:31254064 | ENST00000350437.4 | UCEC | rs1046456 | chr12:31253995 | C/T | 3.01e-05
|












 Gene summary
Gene summary Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez  Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 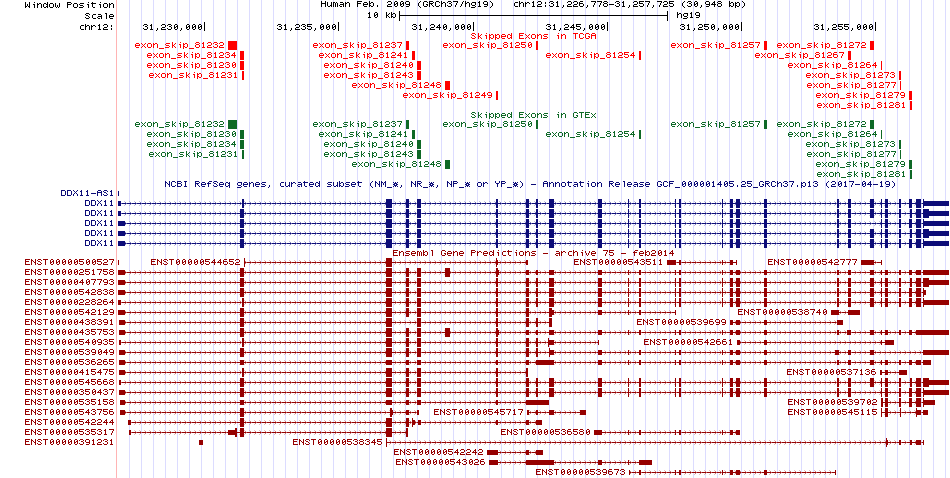
 Expression levels of gene isoforms across TCGA.
Expression levels of gene isoforms across TCGA. Expression levels of gene isoforms across GTEx.
Expression levels of gene isoforms across GTEx. Information of exkip skipping event in TCGA.
Information of exkip skipping event in TCGA. PSI values of skipped exons in TCGA.
PSI values of skipped exons in TCGA. Information of exkip skipping event in GTEx
Information of exkip skipping event in GTEx PSI values of skipped exons in GTEx.
PSI values of skipped exons in GTEx. Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms.
Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms.
Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms. Exon skipping at the protein sequence level and followed lost functional features.
Exon skipping at the protein sequence level and followed lost functional features.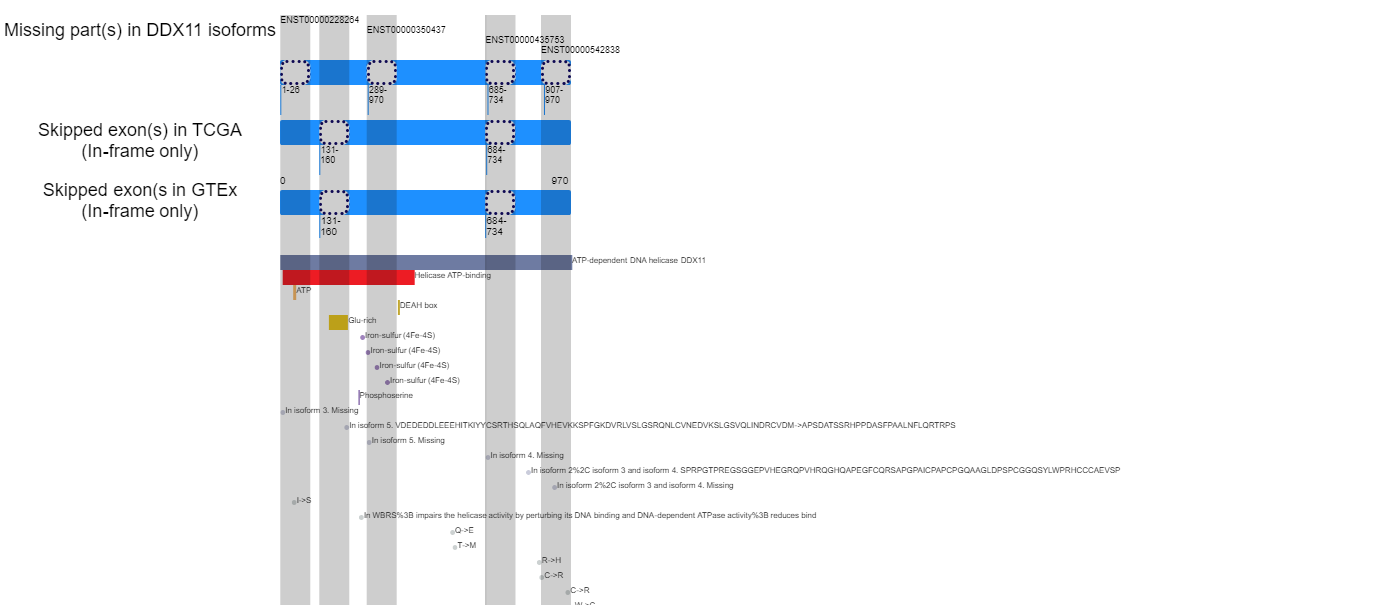
 Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases.
Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases.
Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases. Lost protein functional features of individual exon skipping events in TCGA.
Lost protein functional features of individual exon skipping events in TCGA. Lost protein functional features of individual exon skipping events in GTEx.
Lost protein functional features of individual exon skipping events in GTEx. - Lollipop plot for presenting exon skipping associated SNVs.
- Lollipop plot for presenting exon skipping associated SNVs. - Differential PSIs between mutated versus non-mutated samples.
- Differential PSIs between mutated versus non-mutated samples. - Non-synonymous mutations located in the skipped exons in TCGA.
- Non-synonymous mutations located in the skipped exons in TCGA. - Depth of coverage in the three exons composing exon skipping event
- Depth of coverage in the three exons composing exon skipping event - Non-synonymous mutations located in the skipped exons in CCLE.
- Non-synonymous mutations located in the skipped exons in CCLE. sQTL information located at the skipped exons.
sQTL information located at the skipped exons. Approved drugs targeting this gene.
Approved drugs targeting this gene.  Diseases associated with this gene.
Diseases associated with this gene. 