
|
||||||
|
 | |
 | |
 | |
 | |
 | Open reading frame (ORF) annotation in the exon skipping event |
 | |
 | |
 | Splicing Quantitative Trait Loci (sQTLs) in the skipped exons |
 | Splicing Quantitative Trait Methylation (sQTM) in the skipped exon |
 | |
 |
Gene summary for CNTROB |
 Gene summary Gene summary |
| Gene information | Gene symbol | CNTROB | Gene ID | 116840 |
| Gene name | centrobin, centriole duplication and spindle assembly protein | |
| Synonyms | LIP8|PP1221 | |
| Cytomap | 17p13.1 | |
| Type of gene | protein-coding | |
| Description | centrobinLYST-interacting protein 8LYST-interacting protein LIP8centrobin, centrosomal BRCA2 interacting proteincentrosomal BRCA2-interacting protein | |
| Modification date | 20180523 | |
| UniProtAcc | Q8N137 | |
| Context | PubMed: CNTROB [Title/Abstract] AND exon [Title/Abstract] AND skip [Title/Abstract] - Title (PMID) |
 Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Gene | GO ID | GO term | PubMed ID |
Top |
Exon skipping events across known transcript of Ensembl for CNTROB from UCSC genome browser |
 Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. * Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
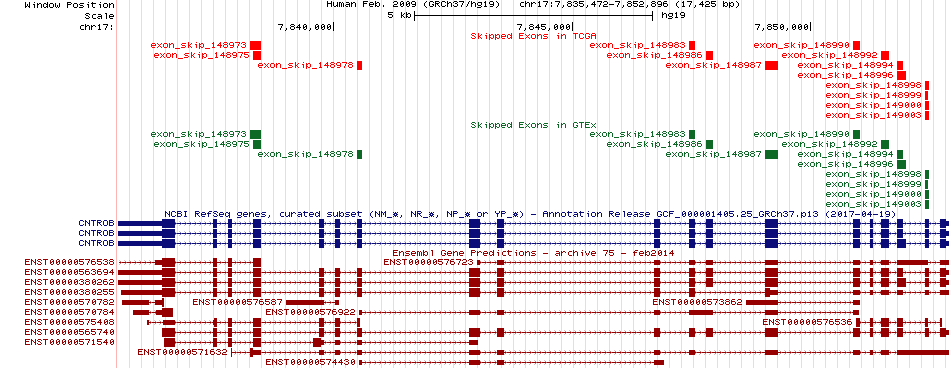 |
Top |
Gene isoform structures and expression levels for CNTROB |
 Expression levels of gene isoforms across TCGA. Expression levels of gene isoforms across TCGA. |
 Expression levels of gene isoforms across GTEx. Expression levels of gene isoforms across GTEx. |
Top |
Exon skipping events with PSIs in TCGA for CNTROB |
 Information of exkip skipping event in TCGA. Information of exkip skipping event in TCGA. |
| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_148973 | 17 | 7837846:7837864:7838236:7838463:7839683:7839800 | 7838236:7838463 | ENSG00000170037.9 | ENST00000571632.1 |
| exon_skip_148975 | 17 | 7837846:7837864:7838306:7838463:7839683:7839800 | 7838306:7838463 | ENSG00000170037.9 | ENST00000380255.3,ENST00000575408.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 |
| exon_skip_148978 | 17 | 7840018:7840135:7840481:7840580:7842830:7843021 | 7840481:7840580 | ENSG00000170037.9 | ENST00000380255.3,ENST00000571540.1,ENST00000571632.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 |
| exon_skip_148983 | 17 | 7846708:7846842:7847440:7847566:7847793:7847956 | 7847440:7847566 | ENSG00000170037.9 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENSG00000170037.9 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENSG00000170037.9 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 |
| exon_skip_148990 | 17 | 7849045:7849304:7850888:7851040:7851233:7851297 | 7850888:7851040 | ENSG00000170037.9 | ENST00000380255.3,ENST00000576723.1,ENST00000571632.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 |
| exon_skip_148992 | 17 | 7851233:7851297:7851474:7851645:7851804:7851933 | 7851474:7851645 | ENSG00000170037.9 | ENST00000380255.3,ENST00000576723.1,ENST00000571632.1,ENST00000576536.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 |
| exon_skip_148994 | 17 | 7851474:7851645:7851804:7851937:7852402:7852455 | 7851804:7851937 | ENSG00000170037.9 | ENST00000380255.3,ENST00000576536.1,ENST00000563694.1 |
| exon_skip_148996 | 17 | 7851474:7851645:7851804:7852003:7852402:7852455 | 7851804:7852003 | ENSG00000170037.9 | ENST00000380262.3 |
| exon_skip_148998 | 17 | 7851804:7851937:7852399:7852475:7852702:7852743 | 7852399:7852475 | ENSG00000170037.9 | ENST00000565740.1 |
| exon_skip_148999 | 17 | 7851804:7851937:7852402:7852455:7852702:7852743 | 7852402:7852455 | ENSG00000170037.9 | ENST00000576536.1 |
| exon_skip_149000 | 17 | 7851804:7851937:7852402:7852475:7852702:7852743 | 7852402:7852475 | ENSG00000170037.9 | ENST00000380255.3,ENST00000563694.1 |
| exon_skip_149003 | 17 | 7851804:7852003:7852402:7852475:7852702:7852743 | 7852402:7852475 | ENSG00000170037.9 | ENST00000380262.3 |
 PSI values of skipped exons in TCGA. PSI values of skipped exons in TCGA. |
Top |
Exon skipping events with PSIs in GTEx for CNTROB |
 Information of exkip skipping event in GTEx Information of exkip skipping event in GTEx |
| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon | ENSG | ENSTs |
| exon_skip_148973 | 17 | 7837846:7837864:7838236:7838463:7839683:7839800 | 7838236:7838463 | ENSG00000170037.9 | ENST00000571632.1 |
| exon_skip_148975 | 17 | 7837846:7837864:7838306:7838463:7839683:7839800 | 7838306:7838463 | ENSG00000170037.9 | ENST00000380262.3,ENST00000563694.1,ENST00000380255.3,ENST00000575408.1,ENST00000565740.1 |
| exon_skip_148978 | 17 | 7840018:7840135:7840481:7840580:7842830:7843021 | 7840481:7840580 | ENSG00000170037.9 | ENST00000380262.3,ENST00000563694.1,ENST00000380255.3,ENST00000565740.1,ENST00000571540.1,ENST00000571632.1 |
| exon_skip_148983 | 17 | 7846708:7846842:7847440:7847566:7847793:7847956 | 7847440:7847566 | ENSG00000170037.9 | ENST00000380262.3,ENST00000563694.1,ENST00000565740.1,ENST00000576723.1 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENSG00000170037.9 | ENST00000380262.3,ENST00000563694.1,ENST00000565740.1,ENST00000576723.1 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENSG00000170037.9 | ENST00000380262.3,ENST00000563694.1,ENST00000565740.1,ENST00000576922.1,ENST00000576723.1 |
| exon_skip_148990 | 17 | 7849045:7849304:7850888:7851040:7851233:7851297 | 7850888:7851040 | ENSG00000170037.9 | ENST00000380262.3,ENST00000563694.1,ENST00000380255.3,ENST00000565740.1,ENST00000571632.1,ENST00000576723.1 |
| exon_skip_148992 | 17 | 7851233:7851297:7851474:7851645:7851804:7851933 | 7851474:7851645 | ENSG00000170037.9 | ENST00000380262.3,ENST00000563694.1,ENST00000380255.3,ENST00000565740.1,ENST00000571632.1,ENST00000576723.1,ENST00000576536.1 |
| exon_skip_148994 | 17 | 7851474:7851645:7851804:7851937:7852402:7852455 | 7851804:7851937 | ENSG00000170037.9 | ENST00000563694.1,ENST00000380255.3,ENST00000576536.1 |
| exon_skip_148996 | 17 | 7851474:7851645:7851804:7852003:7852402:7852455 | 7851804:7852003 | ENSG00000170037.9 | ENST00000380262.3 |
| exon_skip_148998 | 17 | 7851804:7851937:7852399:7852475:7852702:7852743 | 7852399:7852475 | ENSG00000170037.9 | ENST00000565740.1 |
| exon_skip_148999 | 17 | 7851804:7851937:7852402:7852455:7852702:7852743 | 7852402:7852455 | ENSG00000170037.9 | ENST00000576536.1 |
| exon_skip_149000 | 17 | 7851804:7851937:7852402:7852475:7852702:7852743 | 7852402:7852475 | ENSG00000170037.9 | ENST00000563694.1,ENST00000380255.3 |
| exon_skip_149003 | 17 | 7851804:7852003:7852402:7852475:7852702:7852743 | 7852402:7852475 | ENSG00000170037.9 | ENST00000380262.3 |
 PSI values of skipped exons in GTEx. PSI values of skipped exons in GTEx. |
| * Skipped exon sequences. |
Top |
Open reading frame (ORF) annotation in the exon skipping event for CNTROB |
 Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in TCGA based on the Ensembl gene structure combined from isoforms. |
| ENST | Start of skipped exon | End of skipped exon | ORF |
| ENST00000563694 | 7838306 | 7838463 | Frame-shift |
| ENST00000563694 | 7847793 | 7847956 | Frame-shift |
| ENST00000563694 | 7849045 | 7849304 | Frame-shift |
| ENST00000563694 | 7850888 | 7851040 | Frame-shift |
| ENST00000563694 | 7851804 | 7851937 | Frame-shift |
| ENST00000563694 | 7852402 | 7852475 | Frame-shift |
| ENST00000563694 | 7840481 | 7840580 | In-frame |
| ENST00000563694 | 7847440 | 7847566 | In-frame |
| ENST00000563694 | 7851474 | 7851645 | In-frame |
 Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in GTEx based on the Ensembl gene structure combined from isoforms. |
| ENST | Start of skipped exon | End of skipped exon | ORF |
| ENST00000563694 | 7838306 | 7838463 | Frame-shift |
| ENST00000563694 | 7847793 | 7847956 | Frame-shift |
| ENST00000563694 | 7849045 | 7849304 | Frame-shift |
| ENST00000563694 | 7850888 | 7851040 | Frame-shift |
| ENST00000563694 | 7851804 | 7851937 | Frame-shift |
| ENST00000563694 | 7852402 | 7852475 | Frame-shift |
| ENST00000563694 | 7840481 | 7840580 | In-frame |
| ENST00000563694 | 7847440 | 7847566 | In-frame |
| ENST00000563694 | 7851474 | 7851645 | In-frame |
Top |
Infer the effects of exon skipping event on protein functional features for CNTROB |
 Exon skipping at the protein sequence level and followed lost functional features. Exon skipping at the protein sequence level and followed lost functional features.* Click on the image to enlarge it in a new window. |
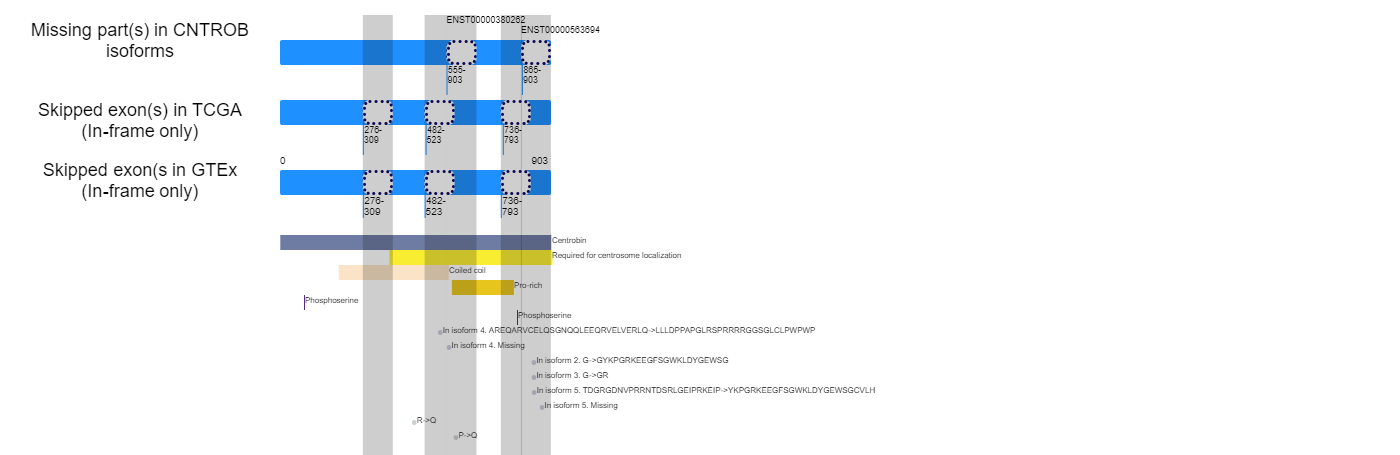 |
 Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in TCGA across genomic, transcript, and protein sequence levels of In-frame cases. |
| ENST | Length of mRNA | Length of AA seq. | Genomic start | Genomic end | mRNA start | mRNA end | AA start | AA end |
| ENST00000563694 | 3722 | 903 | 7840481 | 7840580 | 1754 | 1852 | 276 | 309 |
| ENST00000563694 | 3722 | 903 | 7847440 | 7847566 | 2371 | 2496 | 482 | 523 |
| ENST00000563694 | 3722 | 903 | 7851474 | 7851645 | 3135 | 3305 | 736 | 793 |
 Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in GTEx across genomic, transcript, and protein sequence levels of In-frame cases. |
| ENST | Length of mRNA | Length of AA seq. | Genomic start | Genomic end | mRNA start | mRNA end | AA start | AA end |
| ENST00000563694 | 3722 | 903 | 7840481 | 7840580 | 1754 | 1852 | 276 | 309 |
| ENST00000563694 | 3722 | 903 | 7847440 | 7847566 | 2371 | 2496 | 482 | 523 |
| ENST00000563694 | 3722 | 903 | 7851474 | 7851645 | 3135 | 3305 | 736 | 793 |
 Lost protein functional features of individual exon skipping events in TCGA. Lost protein functional features of individual exon skipping events in TCGA. |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q8N137 | 276 | 309 | 1 | 903 | Chain | ID=PRO_0000076240;Note=Centrobin |
| Q8N137 | 276 | 309 | 196 | 560 | Coiled coil | Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q8N137 | 482 | 523 | 1 | 903 | Chain | ID=PRO_0000076240;Note=Centrobin |
| Q8N137 | 482 | 523 | 196 | 560 | Coiled coil | Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q8N137 | 482 | 523 | 365 | 903 | Region | Note=Required for centrosome localization |
| Q8N137 | 736 | 793 | 555 | 903 | Alternative sequence | ID=VSP_016837;Note=In isoform 4. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:17974005;Dbxref=PMID:17974005 |
| Q8N137 | 736 | 793 | 1 | 903 | Chain | ID=PRO_0000076240;Note=Centrobin |
| Q8N137 | 736 | 793 | 573 | 777 | Compositional bias | Note=Pro-rich |
| Q8N137 | 736 | 793 | 790 | 790 | Modified residue | Note=Phosphoserine;Ontology_term=ECO:0000244;evidence=ECO:0000244|PubMed:23186163;Dbxref=PMID:23186163 |
| Q8N137 | 736 | 793 | 365 | 903 | Region | Note=Required for centrosome localization |
 Lost protein functional features of individual exon skipping events in GTEx. Lost protein functional features of individual exon skipping events in GTEx. |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q8N137 | 276 | 309 | 1 | 903 | Chain | ID=PRO_0000076240;Note=Centrobin |
| Q8N137 | 276 | 309 | 196 | 560 | Coiled coil | Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q8N137 | 482 | 523 | 1 | 903 | Chain | ID=PRO_0000076240;Note=Centrobin |
| Q8N137 | 482 | 523 | 196 | 560 | Coiled coil | Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q8N137 | 482 | 523 | 365 | 903 | Region | Note=Required for centrosome localization |
| Q8N137 | 736 | 793 | 555 | 903 | Alternative sequence | ID=VSP_016837;Note=In isoform 4. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:17974005;Dbxref=PMID:17974005 |
| Q8N137 | 736 | 793 | 1 | 903 | Chain | ID=PRO_0000076240;Note=Centrobin |
| Q8N137 | 736 | 793 | 573 | 777 | Compositional bias | Note=Pro-rich |
| Q8N137 | 736 | 793 | 790 | 790 | Modified residue | Note=Phosphoserine;Ontology_term=ECO:0000244;evidence=ECO:0000244|PubMed:23186163;Dbxref=PMID:23186163 |
| Q8N137 | 736 | 793 | 365 | 903 | Region | Note=Required for centrosome localization |
Top |
SNVs in the skipped exons for CNTROB |
 - Lollipop plot for presenting exon skipping associated SNVs. - Lollipop plot for presenting exon skipping associated SNVs.* Click on the image to enlarge it in a new window. |
 - Differential PSIs between mutated versus non-mutated samples. - Differential PSIs between mutated versus non-mutated samples. |
 - Non-synonymous mutations located in the skipped exons in TCGA. - Non-synonymous mutations located in the skipped exons in TCGA. |
| Cancer type | Sample | ESID | Skipped exon start | Skipped exon end | Mutation start | Mutation end | Mutation type | Reference seq | Mutation seq | AAchange |
| LIHC | TCGA-G3-A3CJ-01 | exon_skip_148990 | 7850889 | 7851040 | 7850891 | 7850891 | Frame_Shift_Del | G | - | p.G666fs |
| LIHC | TCGA-DD-A3A0-01 | exon_skip_148990 | 7850889 | 7851040 | 7850917 | 7850917 | Frame_Shift_Del | C | - | p.F674fs |
| STAD | TCGA-CD-A48C-01 | exon_skip_148992 | 7851475 | 7851645 | 7851480 | 7851483 | Frame_Shift_Del | CTGA | - | p.738_739del |
| STAD | TCGA-CD-A48C-01 | exon_skip_148992 | 7851475 | 7851645 | 7851480 | 7851483 | Frame_Shift_Del | CTGA | - | p.P738fs |
| LIHC | TCGA-RG-A7D4-01 | exon_skip_148994 | 7851805 | 7851937 | 7851880 | 7851898 | Frame_Shift_Del | GGGGGGCTCTGCCTGCTGA | - | p.819_825del |
| LIHC | TCGA-RG-A7D4-01 | exon_skip_148994 | 7851805 | 7851937 | 7851880 | 7851898 | Frame_Shift_Del | GGGGGGCTCTGCCTGCTGA | - | p.WGALPAE819fs |
| LIHC | TCGA-RG-A7D4-01 | exon_skip_148996 | 7851805 | 7852003 | 7851880 | 7851898 | Frame_Shift_Del | GGGGGGCTCTGCCTGCTGA | - | p.819_825del |
| LIHC | TCGA-RG-A7D4-01 | exon_skip_148996 | 7851805 | 7852003 | 7851880 | 7851898 | Frame_Shift_Del | GGGGGGCTCTGCCTGCTGA | - | p.WGALPAE819fs |
| STAD | TCGA-HU-A4GT-01 | exon_skip_148994 | 7851805 | 7851937 | 7851880 | 7851880 | Frame_Shift_Del | G | - | p.W819fs |
| STAD | TCGA-HU-A4GT-01 | exon_skip_148996 | 7851805 | 7852003 | 7851880 | 7851880 | Frame_Shift_Del | G | - | p.W819fs |
| BRCA | TCGA-D8-A1XY-01 | exon_skip_148983 | 7847441 | 7847566 | 7847523 | 7847523 | Nonsense_Mutation | C | T | p.R510* |
| COAD | TCGA-AZ-4615-01 | exon_skip_148983 | 7847441 | 7847566 | 7847523 | 7847523 | Nonsense_Mutation | C | T | p.R510X |
| HNSC | TCGA-CR-7399-01 | exon_skip_148992 | 7851475 | 7851645 | 7851510 | 7851510 | Nonsense_Mutation | C | T | p.Q749* |
| LUSC | TCGA-66-2800-01 | exon_skip_148978 | 7840482 | 7840580 | 7840481 | 7840481 | Splice_Site | G | A | p.T277_splice |
| HNSC | TCGA-D6-6516-01 | exon_skip_148987 | 7849046 | 7849304 | 7849045 | 7849046 | Splice_Site | GG | AA | p.A579_splice |
 - Depth of coverage in the three exons composing exon skipping event - Depth of coverage in the three exons composing exon skipping event |
| Depth of coverage in three exons | Mutation description |
 - Non-synonymous mutations located in the skipped exons in CCLE. - Non-synonymous mutations located in the skipped exons in CCLE. |
| Sample | Skipped exon start | Skipped exon end | Mutation start | Mutation end | Mutation type | Reference seq | Mutation seq | AAchange |
| RH30_SOFT_TISSUE | 7849046 | 7849304 | 7849127 | 7849140 | Frame_Shift_Del | GCCGTGGCCCTGAA | - | p.AVALK606fs |
| LN382_CENTRAL_NERVOUS_SYSTEM | 7849046 | 7849304 | 7849243 | 7849244 | Frame_Shift_Del | TT | - | p.F645fs |
| DND41_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7851805 | 7851937 | 7851880 | 7851880 | Frame_Shift_Del | G | - | p.W819fs |
| DND41_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7851805 | 7852003 | 7851880 | 7851880 | Frame_Shift_Del | G | - | p.W819fs |
| DOV13_OVARY | 7838307 | 7838463 | 7838356 | 7838356 | Missense_Mutation | G | C | p.E163Q |
| DOV13_OVARY | 7838237 | 7838463 | 7838356 | 7838356 | Missense_Mutation | G | C | p.E163Q |
| HS294T_SKIN | 7838307 | 7838463 | 7838356 | 7838356 | Missense_Mutation | G | C | p.E163Q |
| HS294T_SKIN | 7838237 | 7838463 | 7838356 | 7838356 | Missense_Mutation | G | C | p.E163Q |
| HT115_LARGE_INTESTINE | 7838307 | 7838463 | 7838386 | 7838386 | Missense_Mutation | C | T | p.R173W |
| HT115_LARGE_INTESTINE | 7838237 | 7838463 | 7838386 | 7838386 | Missense_Mutation | C | T | p.R173W |
| HSC2_UPPER_AERODIGESTIVE_TRACT | 7838307 | 7838463 | 7838405 | 7838405 | Missense_Mutation | A | G | p.N179S |
| HSC2_UPPER_AERODIGESTIVE_TRACT | 7838237 | 7838463 | 7838405 | 7838405 | Missense_Mutation | A | G | p.N179S |
| HT1376_URINARY_TRACT | 7840482 | 7840580 | 7840507 | 7840507 | Missense_Mutation | T | G | p.L285R |
| CAL851_BREAST | 7840482 | 7840580 | 7840520 | 7840520 | Missense_Mutation | G | A | p.M289I |
| HEC1B_ENDOMETRIUM | 7840482 | 7840580 | 7840525 | 7840525 | Missense_Mutation | C | T | p.A291V |
| HCT15_LARGE_INTESTINE | 7840482 | 7840580 | 7840551 | 7840551 | Missense_Mutation | A | G | p.R300G |
| CMLT1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7840482 | 7840580 | 7840563 | 7840563 | Missense_Mutation | C | T | p.R304W |
| HCC1187_BREAST | 7847794 | 7847956 | 7847801 | 7847801 | Missense_Mutation | C | T | p.R527W |
| HEC59_ENDOMETRIUM | 7847794 | 7847956 | 7847851 | 7847851 | Missense_Mutation | G | T | p.E543D |
| SNU520_STOMACH | 7847794 | 7847956 | 7847859 | 7847859 | Missense_Mutation | G | A | p.R546Q |
| HT144_SKIN | 7847794 | 7847956 | 7847900 | 7847900 | Missense_Mutation | C | T | p.H560Y |
| LS411N_LARGE_INTESTINE | 7847794 | 7847956 | 7847924 | 7847924 | Missense_Mutation | C | A | p.L568I |
| JHH1_LIVER | 7847794 | 7847956 | 7847930 | 7847930 | Missense_Mutation | A | G | p.T570A |
| MEG01_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7849046 | 7849304 | 7849076 | 7849076 | Missense_Mutation | C | T | p.P589S |
| JURKAT_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7849046 | 7849304 | 7849206 | 7849206 | Missense_Mutation | G | A | p.S632N |
| HOP62_LUNG | 7850889 | 7851040 | 7850908 | 7850909 | Missense_Mutation | TG | CT | p.G672W |
| HOP62_LUNG | 7850889 | 7851040 | 7850909 | 7850909 | Missense_Mutation | G | T | p.G672W |
| MELJUSO_SKIN | 7850889 | 7851040 | 7850948 | 7850948 | Missense_Mutation | C | G | p.P685A |
| RL952_ENDOMETRIUM | 7850889 | 7851040 | 7851002 | 7851002 | Missense_Mutation | C | T | p.P703S |
| SNU1040_LARGE_INTESTINE | 7851475 | 7851645 | 7851562 | 7851562 | Missense_Mutation | T | C | p.M766T |
| EN_ENDOMETRIUM | 7851475 | 7851645 | 7851580 | 7851580 | Missense_Mutation | G | T | p.R772L |
| RL952_ENDOMETRIUM | 7851475 | 7851645 | 7851586 | 7851586 | Missense_Mutation | C | T | p.P774L |
| HDLM2_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7851475 | 7851645 | 7851621 | 7851621 | Missense_Mutation | C | T | p.P786S |
| HEC108_ENDOMETRIUM | 7852400 | 7852475 | 7852408 | 7852408 | Missense_Mutation | A | G | p.D840G |
| HEC108_ENDOMETRIUM | 7852403 | 7852455 | 7852408 | 7852408 | Missense_Mutation | A | G | p.D840G |
| HEC108_ENDOMETRIUM | 7852403 | 7852475 | 7852408 | 7852408 | Missense_Mutation | A | G | p.D840G |
| NY_BONE | 7852400 | 7852475 | 7852429 | 7852429 | Missense_Mutation | C | G | p.P847R |
| NY_BONE | 7852403 | 7852455 | 7852429 | 7852429 | Missense_Mutation | C | G | p.P847R |
| NY_BONE | 7852403 | 7852475 | 7852429 | 7852429 | Missense_Mutation | C | G | p.P847R |
| MLMA_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7851805 | 7851937 | 7851858 | 7851858 | Nonsense_Mutation | C | T | p.R812* |
| MLMA_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | 7851805 | 7852003 | 7851858 | 7851858 | Nonsense_Mutation | C | T | p.R812* |
| SNU1040_LARGE_INTESTINE | 7851805 | 7851937 | 7851858 | 7851858 | Nonsense_Mutation | C | T | p.R812* |
| SNU1040_LARGE_INTESTINE | 7851805 | 7852003 | 7851858 | 7851858 | Nonsense_Mutation | C | T | p.R812* |
Top |
Splicing Quantitative Trait Loci (sQTL) in the exon skipping event for CNTROB |
 sQTL information located at the skipped exons. sQTL information located at the skipped exons. |
| Exon skip ID | Chromosome | Three exons | Skippped exon | ENST | Cancer type | SNP id | Location | DNA change (ref/var) | P-value |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | CESC | rs4462665 | chr17:7849087 | C/G | 2.96e-07 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | COAD | rs4462665 | chr17:7849087 | C/G | 5.91e-08 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | HNSC | rs4462665 | chr17:7849087 | C/G | 1.96e-11 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | KIRP | rs4462665 | chr17:7849087 | C/G | 6.53e-16 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | KIRC | rs4462665 | chr17:7849087 | C/G | 1.87e-20 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | LUAD | rs4462665 | chr17:7849087 | C/G | 1.03e-09 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | PCPG | rs4462665 | chr17:7849087 | C/G | 2.83e-12 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | PAAD | rs4462665 | chr17:7849087 | C/G | 1.21e-05 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | PRAD | rs4462665 | chr17:7849087 | C/G | 4.82e-32 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | THYM | rs4462665 | chr17:7849087 | C/G | 3.40e-05 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | TGCT | rs4462665 | chr17:7849087 | C/G | 2.08e-06 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | SKCM | rs4462665 | chr17:7849087 | C/G | 1.87e-05 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | THCA | rs4462665 | chr17:7849087 | C/G | 6.61e-39 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | UVM | rs4462665 | chr17:7849087 | C/G | 5.57e-07 |
| exon_skip_148987 | 17 | 7847793:7847956:7849045:7849304:7850888:7851003 | 7849045:7849304 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000576922.1,ENST00000380262.3 | UCEC | rs4462665 | chr17:7849087 | C/G | 7.39e-05 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | CESC | rs11650083 | chr17:7847955 | C/A | 1.64e-07 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | COAD | rs11650083 | chr17:7847955 | C/A | 2.79e-08 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | HNSC | rs11650083 | chr17:7847955 | C/A | 1.89e-10 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | KIRP | rs11650083 | chr17:7847955 | C/A | 3.91e-18 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | KIRC | rs11650083 | chr17:7847955 | C/A | 2.50e-19 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | LUAD | rs11650083 | chr17:7847955 | C/A | 1.70e-10 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | PCPG | rs11650083 | chr17:7847955 | C/A | 7.33e-13 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | PAAD | rs11650083 | chr17:7847955 | C/A | 2.12e-05 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | PRAD | rs11650083 | chr17:7847955 | C/A | 1.22e-31 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | THYM | rs11650083 | chr17:7847955 | C/A | 4.29e-05 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | TGCT | rs11650083 | chr17:7847955 | C/A | 2.08e-06 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | SKCM | rs11650083 | chr17:7847955 | C/A | 1.87e-05 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | THCA | rs11650083 | chr17:7847955 | C/A | 5.31e-39 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | UVM | rs11650083 | chr17:7847955 | C/A | 5.57e-07 |
| exon_skip_148986 | 17 | 7847440:7847566:7847793:7847956:7849045:7849304 | 7847793:7847956 | ENST00000576723.1,ENST00000565740.1,ENST00000563694.1,ENST00000380262.3 | UCEC | rs11650083 | chr17:7847955 | C/A | 1.04e-04 |
Top |
Splicing Quantitative Trait Methylation (sQTM) in the skipped exon for CNTROB |
Top |
Survival analysis of Splicing Quantitative Trait Methylation (sQTM) in the skipped exon for CNTROB |
Top |
RelatedDrugs for CNTROB |
 Approved drugs targeting this gene. Approved drugs targeting this gene. (DrugBank Version 5.1.0 2018-04-02) |
| Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
RelatedDiseases for CNTROB |
 Diseases associated with this gene. Diseases associated with this gene. (DisGeNet 4.0) |
| Gene | Disease ID | Disease name | # pubmeds | Source |
| CNTROB | C0032460 | Polycystic Ovary Syndrome | 1 | CTD_human |
