|
Home |
Download |
Statistics |
Landscape |
Help |
Contact |
Navigation
1. Exon skipping information for ROSMAP, MSBB, and Mayo2. ORF annotation
3. Analyses of protean functional feature retention
4. sQTM analysis (Splicing Quantitative Trait Loci)
5. Understanding annotation of ExonSkipAD
Search page, example: ASPH
Exon skipped gene search result page
Exon skipped gene annotation result page
1) Gene summary
2) Gene structures and expression levels
3) Exon skipping events with PSIs in RPSMAP, MSBB, and Mayo
4) Open reading frame (ORF) annotation in the exon skipping event
5) Exon skipping events in the canonical protein sequence
6) 3'-UTR located exon skipping events lost miRNA binding sites
7) SNVs in the skipped exons with depth of coverage
8) Correlation with RNA binding proteins (RBPs)
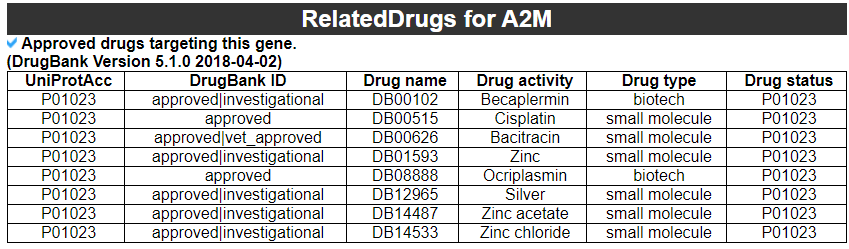
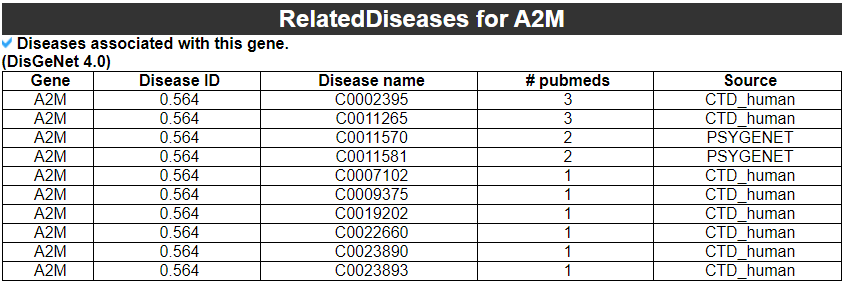
9) Related drugs with this gene
5. Download data and contact us
1. Identifying exon skipping events in ROSMAP, MSBB, and Mayo
We downloaded RNA-Seq bam files and clinical information of three cohorts from the AMP-AD Knowledge Portal. According to the diagnosis annotations of each cohort, we chose AD and control samples per tissue type. We re-mapped RNA-seq bam files to GRCh38 using STAR. To analyze the alternative splicing in AD, we run spladder (2.1.0). We mainly considered exon skipping and mutually exclusive exons.
2. ORF annotation
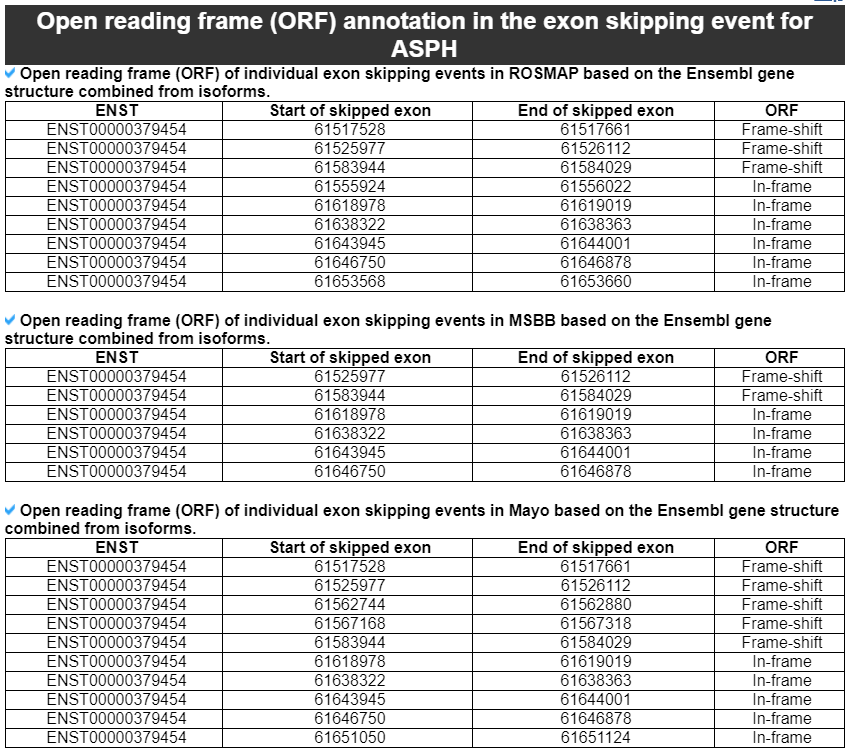
We examined the open reading frames of major isoforms' transcript sequences when there is the specific exon skipping event. When both of the nucleotide start and end positions of exon skipping are located inside of the coding region (CDS) and the number of transcript sequences after exclusion of the skipped exon sequence is a multiple of three, then we reported such exon skipped gene isoform as ‘in-frame’. If there are one or two nucleotide insertions, then we reported such transcript as ‘frame-shift’.
3. Protean features (domains) retention analysis
We searched the retention of 39 protein features of UniProt (6 molecule processing features, 13 region features, the 4 site features, 6 amino acid modification features, 2 natural variation features, 5 experimental info features, and 3 secondary structure features) at the canonical amino acid sequence level with skipped exon loci information. Detailed information about all of the protein features is in UniProt.
4. Understanding ExonSkipAD's annotation categories
Search page, example: ASPH
- Input query: Official HUGO gene symbol or Entrez gene ID.

Exon skipped gene search result page
Select your gene from the gene list.

Exon skipped gene annotation result page
These are ExonSkipAD's annotation categories for your query with links to their corresponding annotation parts.

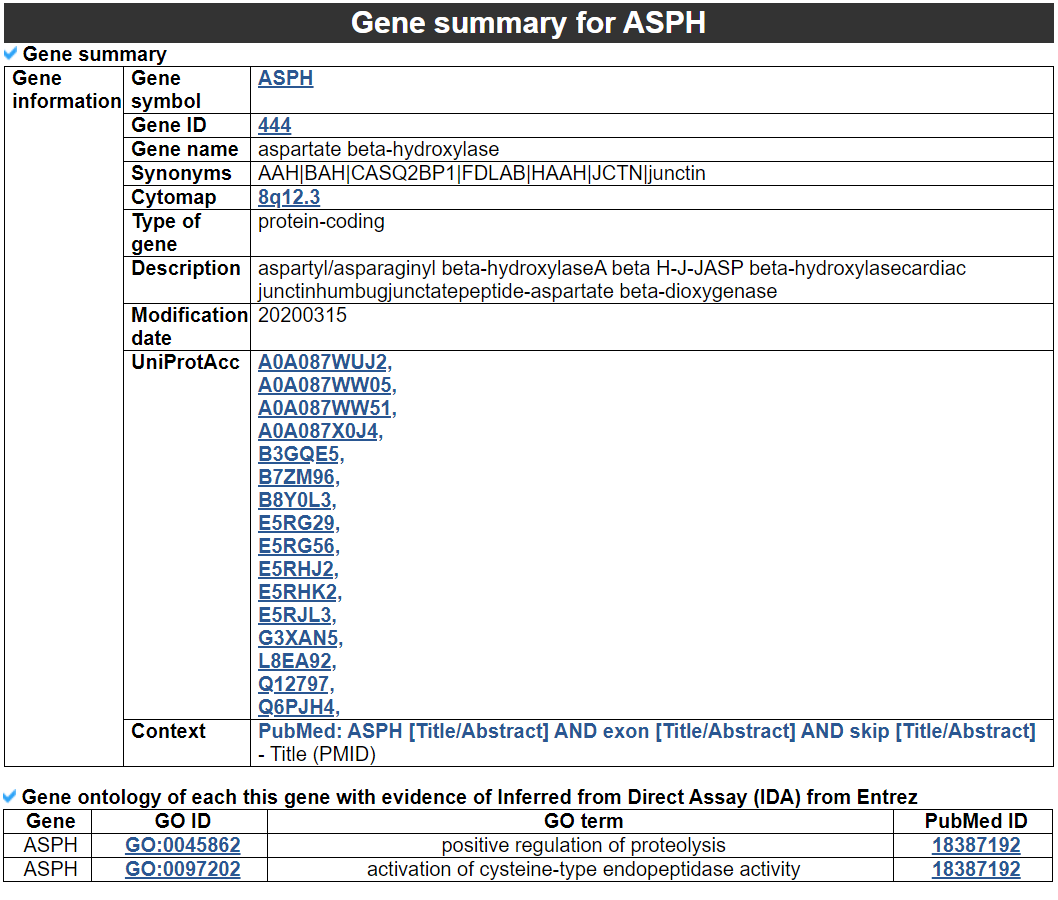
1) Gene summary category
This category shows the information of the exon skipped gene. Firstly, it shows each partner gene's overall information from basic information such as symbol, alias, and locations and ENST accessions. This table also shows the tissue and cancer type information including manually curated PubMed article information.
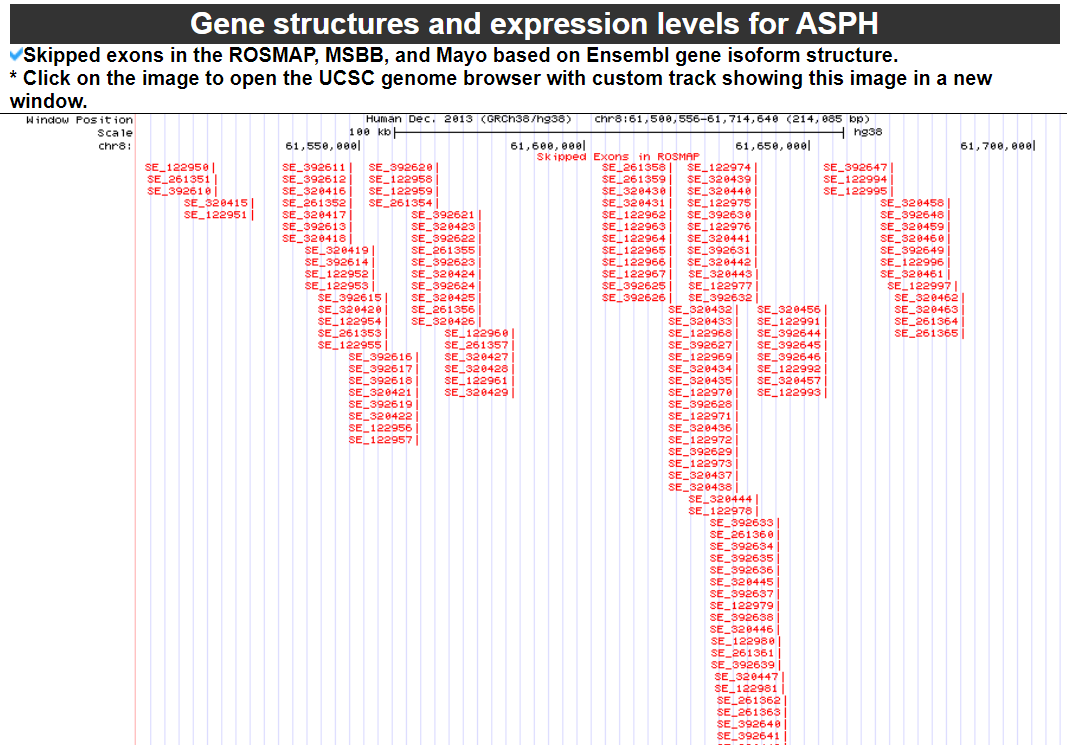
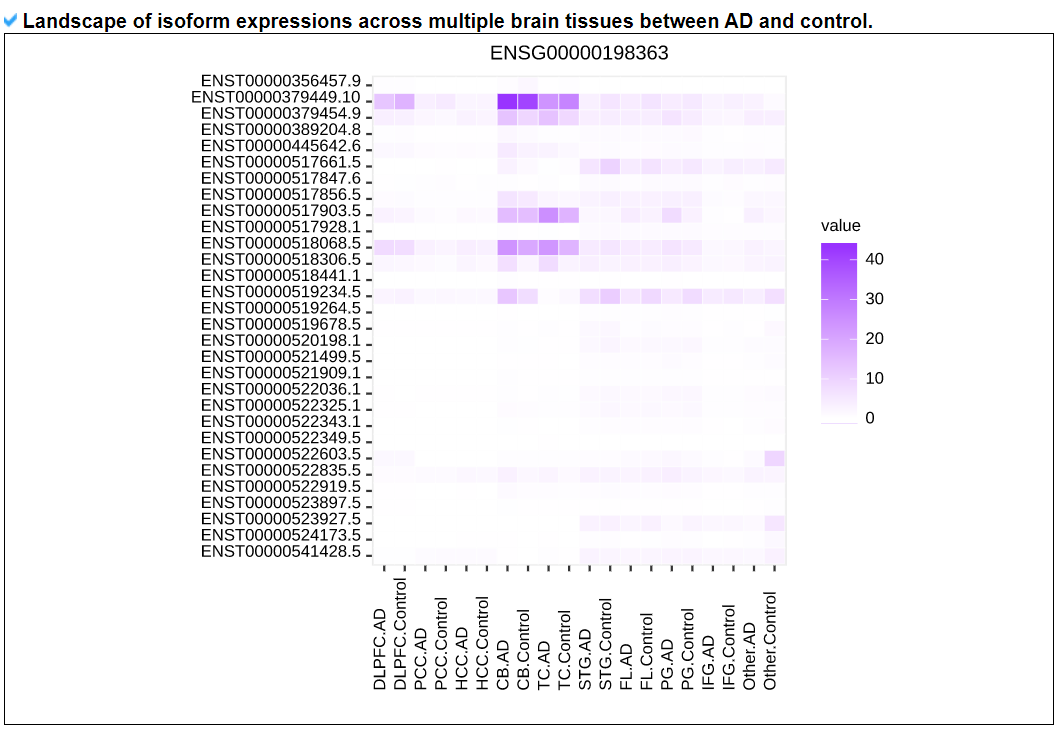
2) 'Gene structures and expression levels' category
This category provides the known gene isoform models based on GENCODE v.32 with expression levels in ROSMAP, MSBB, and Mayo.
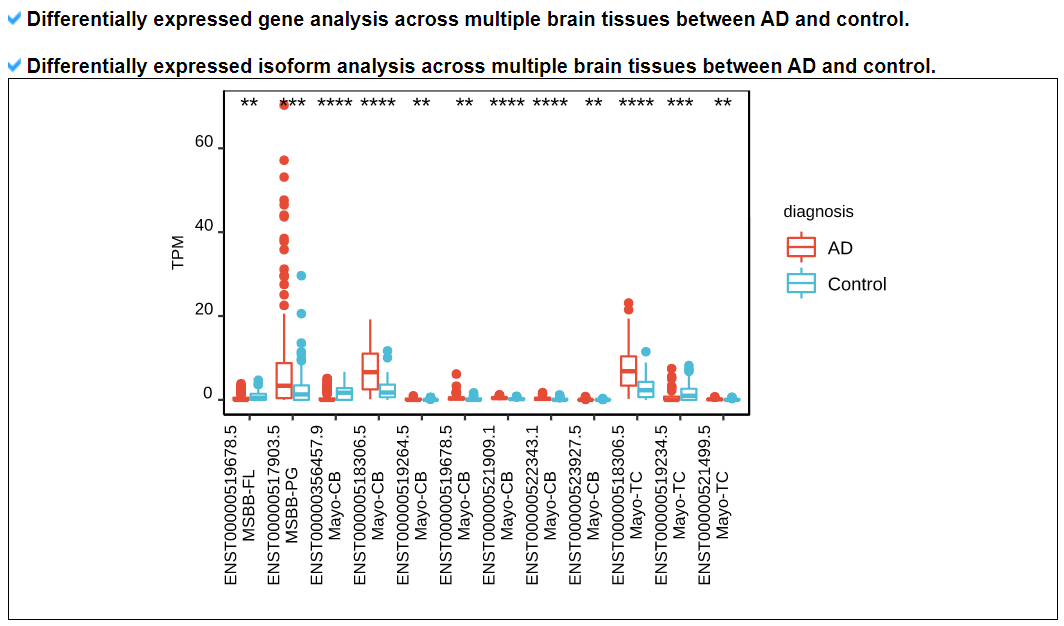
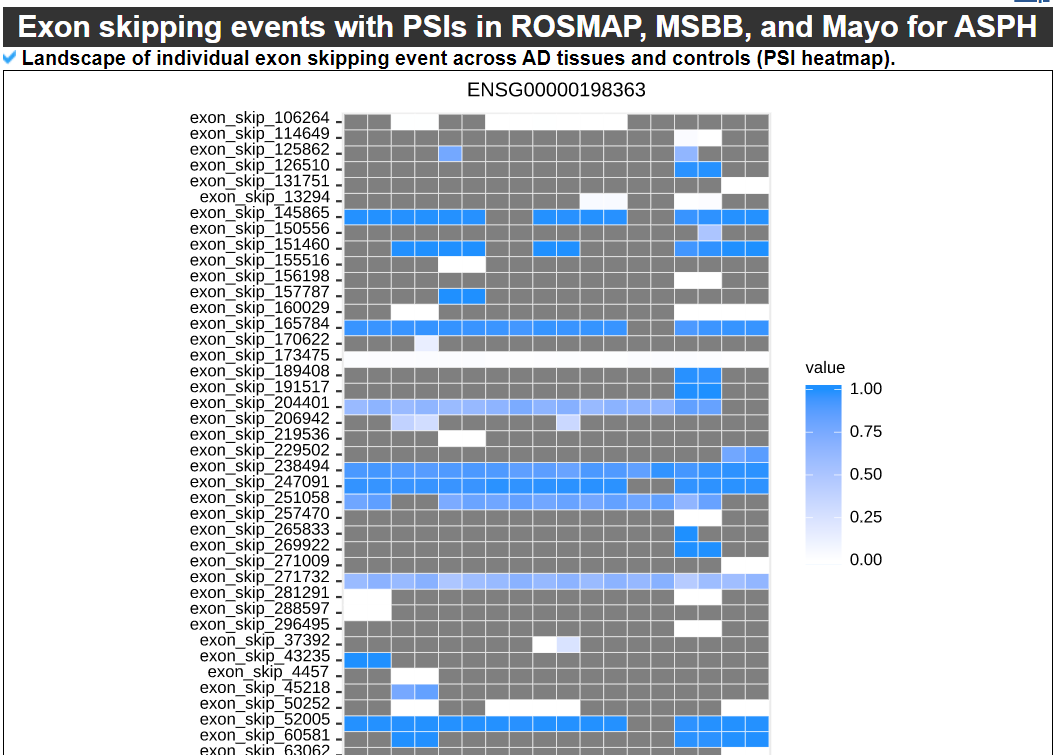
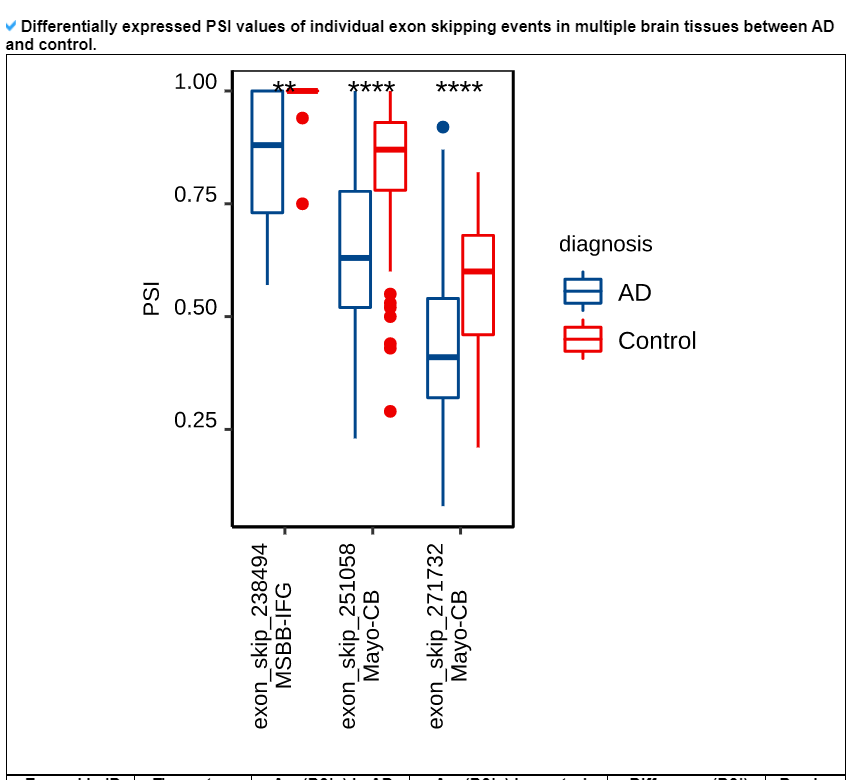
3) 'Exon skipping events with PSIs in ROSMAP, MSBB, and Mayo category
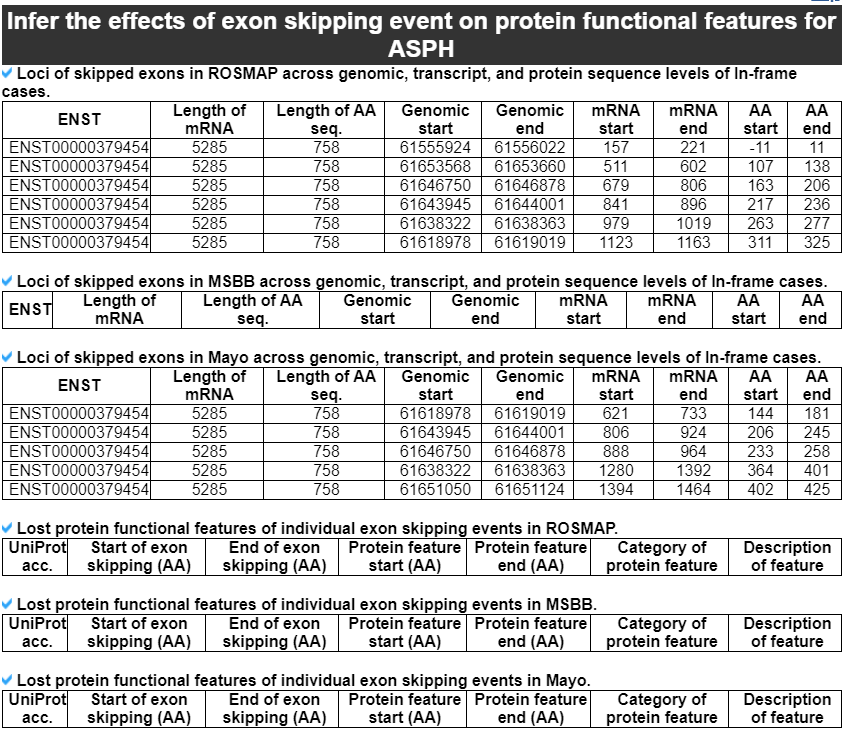
This category provides the detailed exon skipping event information including genomic location with accordant gene isoforms with four splice site matched cases (1st exon's end site, skipped exon's start and end sites, and 3rd exon's start site). Furthermore, this category shows the image of locational map of individual exon skipping events across the canonical gene isoform structure with following the averaged PSI values across 9 brain tissues in three cohorts.

4) 'Open reading frame (ORF) annotation in the exon skipping event' category
This category provides the annotation of the ORF of individual exon skipping events based on the major gene isoform structure.
5) 'Exon skipping events in the canonical protein sequence' category
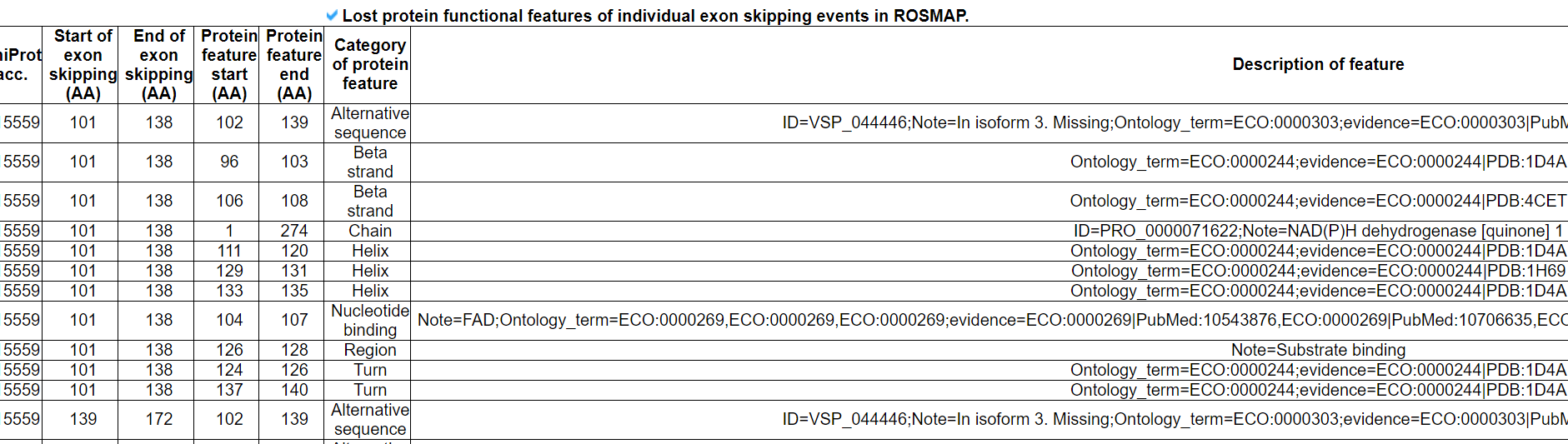
This category provides the detailed information of the protein functional feataures based on the canonical protein sequence. It shows the unique image of presenting skipped exons in the known gene isoforms or novel isoforms in three cohorts. The gray colored background bar through this image is the region of an individual exon skipping event in the canonical protein amino acid sequences. The following table shows the genomic position, transcribed loci, and tranlated loci information of each exon skipping event. From the next table, the user can recognize which functional features were lost due to each exon skipping event. This analysis is done for the in-frame exon skipping events only.
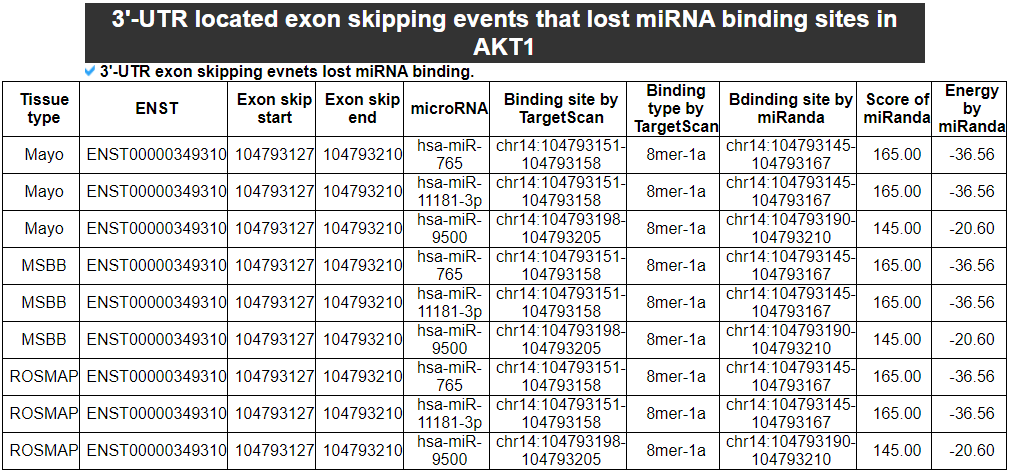
6) '3'-UTR located exon skipping events lost miRNA binding sites' category
This category provides the information of the 3'-UTR located exon skipping events and predicted miRNA binding sites in those. So, the users can expect that these events might lose the regulation of those microRNAs.
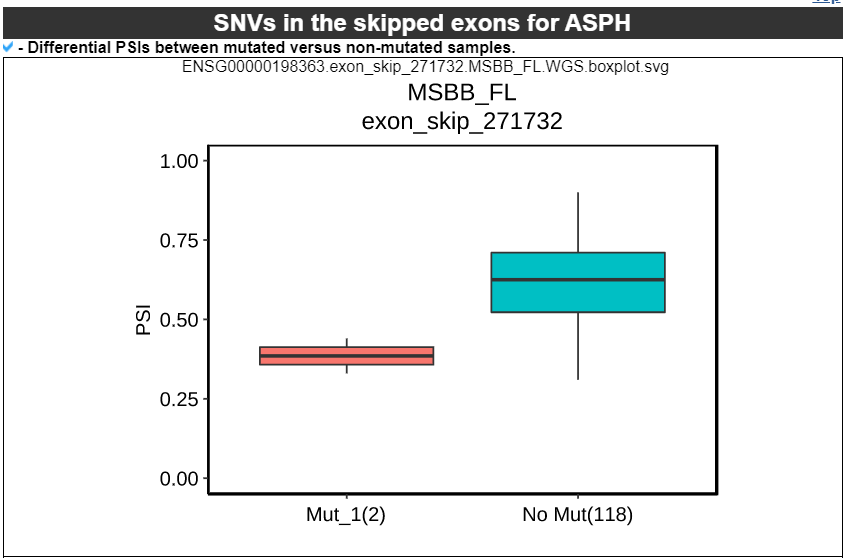
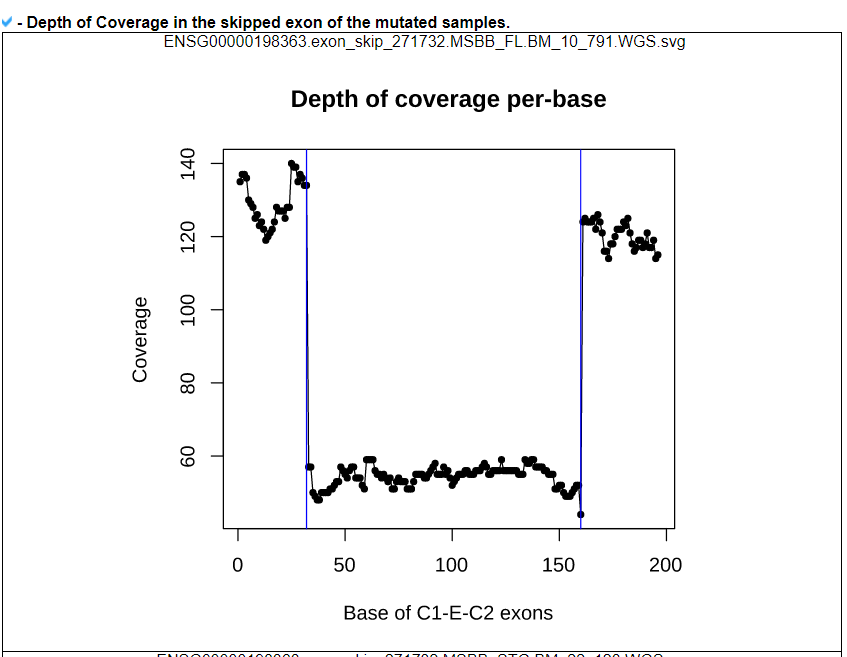
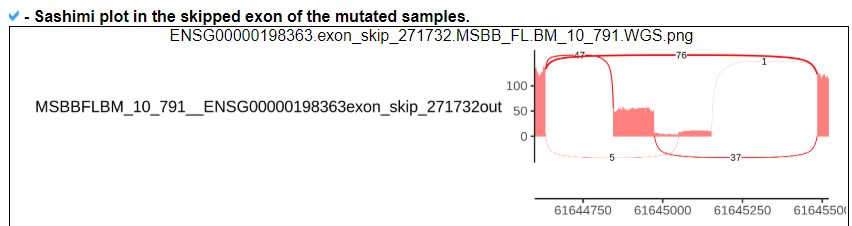
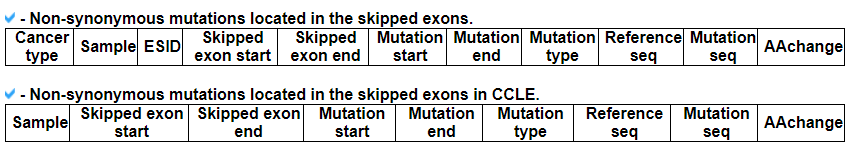
7) 'SNVs in the skipped exons with depth of coverage' category
This category provides the information of mutations associated with exon skipping events. We checked mutations of splice site, nonsense, and frame-shift located in skipped exons. The lollipop plot shows the location on protein domains. With the detailed information of mutation, this category also shows the depth of coverage plot based on three exons, which were involved in the exon skipping event.
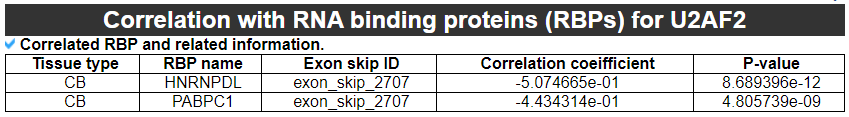
8) 'Correlation with RNA binding proteins (RBPs)' category
This category provides the information about the correlation between RBP expression and the PSI values.
9) 'Related drugs and diseases with this gene category'
This table provides the DrugBank information of approved drugs related with exon skipped gene. Next table provides disease information related to the exon skipped gene.
6. Download data and contact us
Please go to download page and contact page.
