| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 169 | 169 | Natural variant | ID=VAR_069996;Note=In EIEE11. E->G;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:27864847;Dbxref=PMID:23935176,PMID:27864847 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 172 | 172 | Natural variant | ID=VAR_075572;Note=Found in a patient with non-specific acute encephalopathy%3B unknown pathological significance. I->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26311622;Dbxref=dbSNP:rs1376337813,PMID:26311622 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 188 | 188 | Natural variant | ID=VAR_029733;Note=In BFIS3%3B mutant channel inactivates more slowly than wild-type whereas the Na(+) channel conductance is not affected. R->W;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:11371648;Dbxref=dbSNP:rs121917748,PMID:11371648 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 191 | 191 | Natural variant | ID=VAR_078195;Note=Probable disease-associated mutation found in a patient with drug-resistant focal epilepsy. W->G;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519525,PMID:27864847 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 177 | 190 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 156 | 176 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 159 | 201 | 191 | 208 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 278 | 347 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 285 | 285 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 291 | 291 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 297 | 297 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 303 | 303 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 308 | 308 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 236 | 236 | Natural variant | ID=VAR_069999;Note=In EIEE11. T->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1235044536,PMID:23935176 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 240 | 240 | Natural variant | ID=VAR_078455;Note=In BFIS3. A->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 251 | 251 | Natural variant | ID=VAR_078196;Note=In EIEE11. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519528,PMID:27864847 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 252 | 252 | Natural variant | ID=VAR_065176;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs387906687,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 261 | 261 | Natural variant | ID=VAR_065177;Note=In BFIS3%3B increased voltage-gated sodium channel activity%3B faster recovery from inactivation%3B gain of function. V->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:20371507;Dbxref=dbSNP:rs1057520413,PMID:20371507 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_070000;Note=In EIEE11. A->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 263 | 263 | Natural variant | ID=VAR_065178;Note=In EIEE11%3B also found in a patient with neonatal epilepsy with late-onset episodic ataxia%3B increased voltage-gated sodium channel activity%3B increased persistent sodium current%3B gain of function. A->V;Ontology_term=ECO:0000269,EC |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 322 | 322 | Natural variant | ID=VAR_073428;Note=Found in a patient with Dravet syndrome%3B unknown pathological significance. D->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:19783390,ECO:0000269|PubMed:23195492;Dbxref=PMID:19783390,PMID:23195492 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 111 | 456 | Repeat | Note=I;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 232 | 250 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 271 | 369 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 233 | 323 | 251 | 270 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat I;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 850 | 850 | Natural variant | ID=VAR_078459;Note=Found in a patient with schizofrenia%3B unknown pathological significance. R->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 853 | 853 | Natural variant | ID=VAR_070001;Note=In EIEE11%3B phenotype consistent with West syndrome. R->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs794727152,PMID:23935176 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 810 | 823 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 844 | 845 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 790 | 809 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 824 | 843 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 796 | 854 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 854 | 973 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 854 | 973 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 854 | 973 | 910 | 910 | Disulfide bond | Note=Interchain%3B with SCN2B or SCN4B;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:P04775 |
| Q99250 | 854 | 973 | 910 | 910 | Disulfide bond | Note=Interchain%3B with SCN2B or SCN4B;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:P04775 |
| Q99250 | 854 | 973 | 910 | 910 | Disulfide bond | Note=Interchain%3B with SCN2B or SCN4B;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:P04775 |
| Q99250 | 854 | 973 | 910 | 910 | Disulfide bond | Note=Interchain%3B with the conotoxin GVIIJ (when the channel is not linked to SCN2B or SCN4B%3B the bond to SCN2B or SCN4B protects the channel from the inhibition by toxin);Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:P04775 |
| Q99250 | 854 | 973 | 910 | 910 | Disulfide bond | Note=Interchain%3B with the conotoxin GVIIJ (when the channel is not linked to SCN2B or SCN4B%3B the bond to SCN2B or SCN4B protects the channel from the inhibition by toxin);Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:P04775 |
| Q99250 | 854 | 973 | 910 | 910 | Disulfide bond | Note=Interchain%3B with the conotoxin GVIIJ (when the channel is not linked to SCN2B or SCN4B%3B the bond to SCN2B or SCN4B protects the channel from the inhibition by toxin);Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:P04775 |
| Q99250 | 854 | 973 | 950 | 959 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 950 | 959 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 950 | 959 | Disulfide bond | Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 928 | 948 | Intramembrane | Note=Pore-forming;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 928 | 948 | Intramembrane | Note=Pore-forming;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 928 | 948 | Intramembrane | Note=Pore-forming;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 854 | 973 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 854 | 973 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 854 | 973 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 854 | 973 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 854 | 973 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 854 | 973 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 854 | 973 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 854 | 973 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 854 | 973 | 856 | 856 | Natural variant | ID=VAR_078460;Note=In EIEE11. R->L;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 854 | 973 | 856 | 856 | Natural variant | ID=VAR_078460;Note=In EIEE11. R->L;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 854 | 973 | 856 | 856 | Natural variant | ID=VAR_078460;Note=In EIEE11. R->L;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 854 | 973 | 873 | 873 | Natural variant | ID=VAR_078735;Note=In EIEE11. I->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 854 | 973 | 873 | 873 | Natural variant | ID=VAR_078735;Note=In EIEE11. I->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 854 | 973 | 873 | 873 | Natural variant | ID=VAR_078735;Note=In EIEE11. I->M;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 854 | 973 | 876 | 876 | Natural variant | ID=VAR_070002;Note=In EIEE11%3B the disease progresses to West syndrome. N->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 854 | 973 | 876 | 876 | Natural variant | ID=VAR_070002;Note=In EIEE11%3B the disease progresses to West syndrome. N->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 854 | 973 | 876 | 876 | Natural variant | ID=VAR_070002;Note=In EIEE11%3B the disease progresses to West syndrome. N->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=PMID:23935176 |
| Q99250 | 854 | 973 | 892 | 892 | Natural variant | ID=VAR_029737;Note=In BFIS3. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917751,PMID:15048894 |
| Q99250 | 854 | 973 | 892 | 892 | Natural variant | ID=VAR_029737;Note=In BFIS3. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917751,PMID:15048894 |
| Q99250 | 854 | 973 | 892 | 892 | Natural variant | ID=VAR_029737;Note=In BFIS3. V->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917751,PMID:15048894 |
| Q99250 | 854 | 973 | 896 | 896 | Natural variant | ID=VAR_078197;Note=In EIEE11. A->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519526,PMID:27864847 |
| Q99250 | 854 | 973 | 896 | 896 | Natural variant | ID=VAR_078197;Note=In EIEE11. A->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519526,PMID:27864847 |
| Q99250 | 854 | 973 | 896 | 896 | Natural variant | ID=VAR_078197;Note=In EIEE11. A->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs1057519526,PMID:27864847 |
| Q99250 | 854 | 973 | 905 | 905 | Natural variant | ID=VAR_078461;Note=In EIEE11. K->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23708187,ECO:0000269|PubMed:26291284;Dbxref=dbSNP:rs796053119,PMID:23708187,PMID:26291284 |
| Q99250 | 854 | 973 | 905 | 905 | Natural variant | ID=VAR_078461;Note=In EIEE11. K->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23708187,ECO:0000269|PubMed:26291284;Dbxref=dbSNP:rs796053119,PMID:23708187,PMID:26291284 |
| Q99250 | 854 | 973 | 905 | 905 | Natural variant | ID=VAR_078461;Note=In EIEE11. K->N;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23708187,ECO:0000269|PubMed:26291284;Dbxref=dbSNP:rs796053119,PMID:23708187,PMID:26291284 |
| Q99250 | 854 | 973 | 908 | 908 | Natural variant | ID=VAR_078462;Note=Found in a patient with schizofrenia%3B unknown pathological significance. K->R;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=dbSNP:rs2228980,PMID:26555645 |
| Q99250 | 854 | 973 | 908 | 908 | Natural variant | ID=VAR_078462;Note=Found in a patient with schizofrenia%3B unknown pathological significance. K->R;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=dbSNP:rs2228980,PMID:26555645 |
| Q99250 | 854 | 973 | 908 | 908 | Natural variant | ID=VAR_078462;Note=Found in a patient with schizofrenia%3B unknown pathological significance. K->R;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=dbSNP:rs2228980,PMID:26555645 |
| Q99250 | 854 | 973 | 928 | 928 | Natural variant | ID=VAR_078463;Note=In EIEE11%3B mild form with ataxia. F->C;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23708187,ECO:0000269|PubMed:26291284;Dbxref=PMID:23708187,PMID:26291284 |
| Q99250 | 854 | 973 | 928 | 928 | Natural variant | ID=VAR_078463;Note=In EIEE11%3B mild form with ataxia. F->C;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23708187,ECO:0000269|PubMed:26291284;Dbxref=PMID:23708187,PMID:26291284 |
| Q99250 | 854 | 973 | 928 | 928 | Natural variant | ID=VAR_078463;Note=In EIEE11%3B mild form with ataxia. F->C;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23708187,ECO:0000269|PubMed:26291284;Dbxref=PMID:23708187,PMID:26291284 |
| Q99250 | 854 | 973 | 937 | 937 | Natural variant | ID=VAR_078464;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. R->C;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=dbSNP:r |
| Q99250 | 854 | 973 | 937 | 937 | Natural variant | ID=VAR_078464;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. R->C;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=dbSNP:r |
| Q99250 | 854 | 973 | 937 | 937 | Natural variant | ID=VAR_078464;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. R->C;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=dbSNP:r |
| Q99250 | 854 | 973 | 937 | 937 | Natural variant | ID=VAR_078465;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. R->H;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID:28 |
| Q99250 | 854 | 973 | 937 | 937 | Natural variant | ID=VAR_078465;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. R->H;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID:28 |
| Q99250 | 854 | 973 | 937 | 937 | Natural variant | ID=VAR_078465;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. R->H;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID:28 |
| Q99250 | 854 | 973 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 854 | 973 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 854 | 973 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 854 | 973 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 864 | 879 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 864 | 879 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 864 | 879 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 899 | 927 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 899 | 927 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 899 | 927 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 949 | 961 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 949 | 961 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 949 | 961 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 854 | 973 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 846 | 863 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 880 | 898 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 880 | 898 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 880 | 898 | Transmembrane | Note=Helical%3B Name%3DS5 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 854 | 973 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 1114 | 1116 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:4RLY |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 976 | 976 | Natural variant | ID=VAR_078467;Note=Found in a patient with autism%3B unknown pathological significance. N->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26291284;Dbxref=PMID:26291284 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 987 | 987 | Natural variant | ID=VAR_078736;Note=In EIEE11. S->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053124,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_070003;Note=In EIEE11%3B the disease progresses to West syndrome. E->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:23935176,ECO:0000269|PubMed:26993267;Dbxref=dbSNP:rs796053126,PMID:23935176,PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 999 | 999 | Natural variant | ID=VAR_078737;Note=In EIEE11. E->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1001 | 1001 | Natural variant | ID=VAR_078468;Note=In BFIS3. N->K;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:16417554,ECO:0000269|PubMed:23360469;Dbxref=PMID:16417554,PMID:23360469 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1003 | 1003 | Natural variant | ID=VAR_029738;Note=In BFIS3. L->I;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15048894;Dbxref=dbSNP:rs121917754,PMID:15048894 |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 1128 | 1128 | Natural variant | ID=VAR_078470;Note=Found in a patient with acute encephalitis with refractory and repetitive partial seizures%3B unknown pathological significance. M->T;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:22591750;Dbxref=dbSNP:rs373780066,PMID:22591750 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 741 | 1013 | Repeat | Note=II;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 983 | 1209 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 973 | 1133 | 962 | 982 | Transmembrane | Note=Helical%3B Name%3DS6 of repeat II;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1225 | 1283 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1225 | 1283 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1225 | 1283 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1225 | 1283 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1225 | 1283 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1225 | 1283 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1225 | 1283 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1225 | 1283 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1225 | 1283 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1225 | 1283 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1225 | 1283 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1225 | 1283 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1225 | 1283 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1225 | 1283 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1225 | 1283 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1225 | 1283 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1225 | 1283 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1225 | 1283 | 1260 | 1260 | Natural variant | ID=VAR_078738;Note=In EIEE11. K->E;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1225 | 1283 | 1260 | 1260 | Natural variant | ID=VAR_078738;Note=In EIEE11. K->E;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1225 | 1283 | 1260 | 1260 | Natural variant | ID=VAR_078738;Note=In EIEE11. K->E;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1225 | 1283 | 1260 | 1260 | Natural variant | ID=VAR_078739;Note=In EIEE11. K->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1225 | 1283 | 1260 | 1260 | Natural variant | ID=VAR_078739;Note=In EIEE11. K->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1225 | 1283 | 1260 | 1260 | Natural variant | ID=VAR_078739;Note=In EIEE11. K->Q;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1225 | 1283 | 1282 | 1282 | Natural variant | ID=VAR_078471;Note=Found in a patient with schizofrenia%3B unknown pathological significance. V->F;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=dbSNP:rs1184922927,PMID:26555645 |
| Q99250 | 1225 | 1283 | 1282 | 1282 | Natural variant | ID=VAR_078471;Note=Found in a patient with schizofrenia%3B unknown pathological significance. V->F;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=dbSNP:rs1184922927,PMID:26555645 |
| Q99250 | 1225 | 1283 | 1282 | 1282 | Natural variant | ID=VAR_078471;Note=Found in a patient with schizofrenia%3B unknown pathological significance. V->F;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=dbSNP:rs1184922927,PMID:26555645 |
| Q99250 | 1225 | 1283 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1228 | 1240 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1228 | 1240 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1228 | 1240 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1260 | 1273 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1260 | 1273 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1260 | 1273 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1225 | 1283 | 1210 | 1227 | Transmembrane | Note=Helical%3B Name%3DS1 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1210 | 1227 | Transmembrane | Note=Helical%3B Name%3DS1 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1210 | 1227 | Transmembrane | Note=Helical%3B Name%3DS1 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1241 | 1259 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1241 | 1259 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1241 | 1259 | Transmembrane | Note=Helical%3B Name%3DS2 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1274 | 1292 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1274 | 1292 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1225 | 1283 | 1274 | 1292 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1283 | 1324 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1283 | 1324 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1283 | 1324 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1283 | 1324 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1283 | 1324 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1283 | 1324 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1283 | 1324 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1283 | 1324 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1283 | 1324 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1283 | 1324 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1283 | 1324 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1283 | 1324 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1283 | 1324 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1283 | 1324 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1283 | 1324 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1283 | 1324 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1283 | 1324 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1283 | 1324 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1283 | 1324 | 1312 | 1312 | Natural variant | ID=VAR_073429;Note=In EIEE11%3B modified voltage-gated sodium channel activity%3B activated with lowered voltage sensitivity%3B disturbed fast and slow inactivation. R->T;Ontology_term=ECO:0000269,ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:197833 |
| Q99250 | 1283 | 1324 | 1312 | 1312 | Natural variant | ID=VAR_073429;Note=In EIEE11%3B modified voltage-gated sodium channel activity%3B activated with lowered voltage sensitivity%3B disturbed fast and slow inactivation. R->T;Ontology_term=ECO:0000269,ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:197833 |
| Q99250 | 1283 | 1324 | 1312 | 1312 | Natural variant | ID=VAR_073429;Note=In EIEE11%3B modified voltage-gated sodium channel activity%3B activated with lowered voltage sensitivity%3B disturbed fast and slow inactivation. R->T;Ontology_term=ECO:0000269,ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:197833 |
| Q99250 | 1283 | 1324 | 1316 | 1316 | Natural variant | ID=VAR_078198;Note=In EIEE11. A->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs796053130,PMID:27864847 |
| Q99250 | 1283 | 1324 | 1316 | 1316 | Natural variant | ID=VAR_078198;Note=In EIEE11. A->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs796053130,PMID:27864847 |
| Q99250 | 1283 | 1324 | 1316 | 1316 | Natural variant | ID=VAR_078198;Note=In EIEE11. A->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:27864847;Dbxref=dbSNP:rs796053130,PMID:27864847 |
| Q99250 | 1283 | 1324 | 1319 | 1319 | Natural variant | ID=VAR_029739;Note=In BFIS3%3B modified voltage-gated sodium channel activity%3B modified voltage dependence of activation and inactivation. R->Q;Ontology_term=ECO:0000269,ECO:0000269,ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:15048894,ECO:000026 |
| Q99250 | 1283 | 1324 | 1319 | 1319 | Natural variant | ID=VAR_029739;Note=In BFIS3%3B modified voltage-gated sodium channel activity%3B modified voltage dependence of activation and inactivation. R->Q;Ontology_term=ECO:0000269,ECO:0000269,ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:15048894,ECO:000026 |
| Q99250 | 1283 | 1324 | 1319 | 1319 | Natural variant | ID=VAR_029739;Note=In BFIS3%3B modified voltage-gated sodium channel activity%3B modified voltage dependence of activation and inactivation. R->Q;Ontology_term=ECO:0000269,ECO:0000269,ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:15048894,ECO:000026 |
| Q99250 | 1283 | 1324 | 1321 | 1321 | Natural variant | ID=VAR_078740;Note=In BFIS3%3B unknown pathological significance. E->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25982755;Dbxref=PMID:25982755 |
| Q99250 | 1283 | 1324 | 1321 | 1321 | Natural variant | ID=VAR_078740;Note=In BFIS3%3B unknown pathological significance. E->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25982755;Dbxref=PMID:25982755 |
| Q99250 | 1283 | 1324 | 1321 | 1321 | Natural variant | ID=VAR_078740;Note=In BFIS3%3B unknown pathological significance. E->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25982755;Dbxref=PMID:25982755 |
| Q99250 | 1283 | 1324 | 1323 | 1323 | Natural variant | ID=VAR_070004;Note=In EIEE11%3B the disease progresses to West syndrome. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1057519523,PMID:23935176 |
| Q99250 | 1283 | 1324 | 1323 | 1323 | Natural variant | ID=VAR_070004;Note=In EIEE11%3B the disease progresses to West syndrome. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1057519523,PMID:23935176 |
| Q99250 | 1283 | 1324 | 1323 | 1323 | Natural variant | ID=VAR_070004;Note=In EIEE11%3B the disease progresses to West syndrome. M->V;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23935176;Dbxref=dbSNP:rs1057519523,PMID:23935176 |
| Q99250 | 1283 | 1324 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1293 | 1300 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1293 | 1300 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1293 | 1300 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1320 | 1336 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1320 | 1336 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1320 | 1336 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1283 | 1324 | 1274 | 1292 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1283 | 1324 | 1274 | 1292 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1283 | 1324 | 1274 | 1292 | Transmembrane | Note=Helical%3B Name%3DS3 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1283 | 1324 | 1301 | 1319 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1283 | 1324 | 1301 | 1319 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1283 | 1324 | 1301 | 1319 | Transmembrane | Note=Helical%3B Name%3DS4 of repeat III;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1418 | 1436 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1418 | 1436 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1418 | 1436 | 1 | 2005 | Chain | ID=PRO_0000048491;Note=Sodium channel protein type 2 subunit alpha |
| Q99250 | 1418 | 1436 | 1409 | 1430 | Intramembrane | Note=Pore-forming;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1418 | 1436 | 1409 | 1430 | Intramembrane | Note=Pore-forming;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1418 | 1436 | 1409 | 1430 | Intramembrane | Note=Pore-forming;Ontology_term=ECO:0000250;evidence=ECO:0000250|UniProtKB:D0E0C2 |
| Q99250 | 1418 | 1436 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1418 | 1436 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1418 | 1436 | 102 | 2005 | Natural variant | ID=VAR_078450;Note=Probable disease-associated mutation found in a patient with intractable epilepsy and severe mental decline%3B non-conducting%3B loss of voltage-gated sodium channel activity%3B dominant-negatif. Missing;Ontology_term=ECO:0000269;eviden |
| Q99250 | 1418 | 1436 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1418 | 1436 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1418 | 1436 | 169 | 2005 | Natural variant | ID=VAR_078453;Note=Found in a patient with schizofrenia%3B unknown pathological significance. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26555645;Dbxref=PMID:26555645 |
| Q99250 | 1418 | 1436 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1418 | 1436 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1418 | 1436 | 583 | 2005 | Natural variant | ID=VAR_078732;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:25969726;Dbxref=PMID:25969726 |
| Q99250 | 1418 | 1436 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1418 | 1436 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1418 | 1436 | 959 | 2005 | Natural variant | ID=VAR_078466;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1418 | 1436 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1418 | 1436 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1418 | 1436 | 1013 | 2005 | Natural variant | ID=VAR_078469;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B loss of voltage-gated sodium channel activity%3B non-conducting. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:28256214;Dbxref=PMID |
| Q99250 | 1418 | 1436 | 1398 | 2005 | Natural variant | ID=VAR_078742;Note=In EIEE11. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23033978;Dbxref=PMID:23033978 |
| Q99250 | 1418 | 1436 | 1398 | 2005 | Natural variant | ID=VAR_078742;Note=In EIEE11. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23033978;Dbxref=PMID:23033978 |
| Q99250 | 1418 | 1436 | 1398 | 2005 | Natural variant | ID=VAR_078742;Note=In EIEE11. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23033978;Dbxref=PMID:23033978 |
| Q99250 | 1418 | 1436 | 1420 | 1420 | Natural variant | ID=VAR_078475;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B decreased voltage-gated sodium channel activity%3B faster channel inactivation%3B fewer channels contribution to macroscopic currents and fewer cha |
| Q99250 | 1418 | 1436 | 1420 | 1420 | Natural variant | ID=VAR_078475;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B decreased voltage-gated sodium channel activity%3B faster channel inactivation%3B fewer channels contribution to macroscopic currents and fewer cha |
| Q99250 | 1418 | 1436 | 1420 | 1420 | Natural variant | ID=VAR_078475;Note=Probable disease-associated mutation found in a patient with autism spectrum disorder%3B decreased voltage-gated sodium channel activity%3B faster channel inactivation%3B fewer channels contribution to macroscopic currents and fewer cha |
| Q99250 | 1418 | 1436 | 1422 | 1422 | Natural variant | ID=VAR_070008;Note=Probable disease-associated mutation found in a boy with infantile spasms and bitemporal glucose hypometabolism. K->E;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23827426;Dbxref=dbSNP:rs796053137,PMID:23827426 |
| Q99250 | 1418 | 1436 | 1422 | 1422 | Natural variant | ID=VAR_070008;Note=Probable disease-associated mutation found in a boy with infantile spasms and bitemporal glucose hypometabolism. K->E;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23827426;Dbxref=dbSNP:rs796053137,PMID:23827426 |
| Q99250 | 1418 | 1436 | 1422 | 1422 | Natural variant | ID=VAR_070008;Note=Probable disease-associated mutation found in a boy with infantile spasms and bitemporal glucose hypometabolism. K->E;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:23827426;Dbxref=dbSNP:rs796053137,PMID:23827426 |
| Q99250 | 1418 | 1436 | 1435 | 2005 | Natural variant | ID=VAR_078743;Note=In EIEE11. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1418 | 1436 | 1435 | 2005 | Natural variant | ID=VAR_078743;Note=In EIEE11. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1418 | 1436 | 1435 | 2005 | Natural variant | ID=VAR_078743;Note=In EIEE11. Missing;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:26993267;Dbxref=PMID:26993267 |
| Q99250 | 1418 | 1436 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1418 | 1436 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1418 | 1436 | 1190 | 1504 | Repeat | Note=III;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1418 | 1436 | 1431 | 1447 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1418 | 1436 | 1431 | 1447 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q99250 | 1418 | 1436 | 1431 | 1447 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000305;evidence=ECO:0000305 |












 Gene summary
Gene summary Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez  Skipped exons in the ROSMAP, MSBB, and Mayo based on Ensembl gene isoform structure.
Skipped exons in the ROSMAP, MSBB, and Mayo based on Ensembl gene isoform structure. 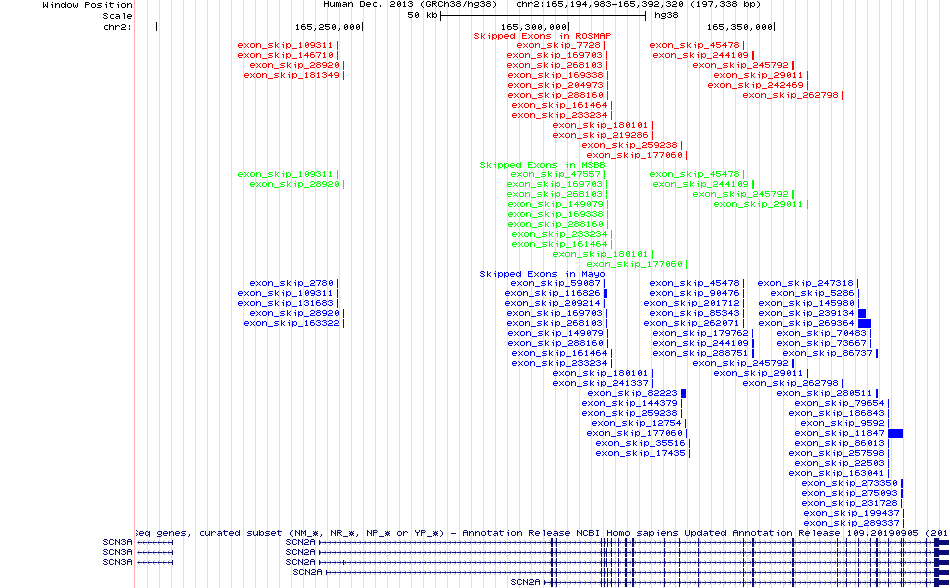
 Differentially expressed gene analysis across multiple brain tissues between AD and control.
Differentially expressed gene analysis across multiple brain tissues between AD and control. Differentially expressed isoform analysis across multiple brain tissues between AD and control.
Differentially expressed isoform analysis across multiple brain tissues between AD and control. Landscape of isoform expressions across multiple brain tissues between AD and control.
Landscape of isoform expressions across multiple brain tissues between AD and control. Landscape of individual exon skipping event across AD tissues and controls (PSI heatmap).
Landscape of individual exon skipping event across AD tissues and controls (PSI heatmap). All exon skipping events in AD cohorts.
All exon skipping events in AD cohorts. Differentially expressed PSI values of individual exon skipping events in multiple brain tissues between AD and control.
Differentially expressed PSI values of individual exon skipping events in multiple brain tissues between AD and control. Open reading frame (ORF) of individual exon skipping events in ROSMAP based on the Ensembl gene structure combined from isoforms.
Open reading frame (ORF) of individual exon skipping events in ROSMAP based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in MSBB based on the Ensembl gene structure combined from isoforms.
Open reading frame (ORF) of individual exon skipping events in MSBB based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in Mayo based on the Ensembl gene structure combined from isoforms.
Open reading frame (ORF) of individual exon skipping events in Mayo based on the Ensembl gene structure combined from isoforms.
 Loci of skipped exons in ROSMAP across genomic, transcript, and protein sequence levels of In-frame cases.
Loci of skipped exons in ROSMAP across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in MSBB across genomic, transcript, and protein sequence levels of In-frame cases.
Loci of skipped exons in MSBB across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in Mayo across genomic, transcript, and protein sequence levels of In-frame cases.
Loci of skipped exons in Mayo across genomic, transcript, and protein sequence levels of In-frame cases. Lost protein functional features of individual exon skipping events in ROSMAP.
Lost protein functional features of individual exon skipping events in ROSMAP. Lost protein functional features of individual exon skipping events in MSBB.
Lost protein functional features of individual exon skipping events in MSBB. Lost protein functional features of individual exon skipping events in Mayo.
Lost protein functional features of individual exon skipping events in Mayo. 3'-UTR exon skipping evnets lost miRNA binding.
3'-UTR exon skipping evnets lost miRNA binding. - Differential PSIs between mutated versus non-mutated samples.
- Differential PSIs between mutated versus non-mutated samples. - Depth of Coverage in the skipped exon of the mutated samples.
- Depth of Coverage in the skipped exon of the mutated samples. - Sashimi plot in the skipped exon of the mutated samples.
- Sashimi plot in the skipped exon of the mutated samples. - Non-synonymous mutations located in the skipped exons.
- Non-synonymous mutations located in the skipped exons. - Non-synonymous mutations located in the skipped exons in CCLE.
- Non-synonymous mutations located in the skipped exons in CCLE. Associated exon skipping events with Braak staging or Clinical Dementia Rating (CDR).
Associated exon skipping events with Braak staging or Clinical Dementia Rating (CDR). sQTL information located at the skipped exons.
sQTL information located at the skipped exons. Correlated RBP and related information.
Correlated RBP and related information. Approved drugs targeting this gene.
Approved drugs targeting this gene.  Diseases associated with this gene.
Diseases associated with this gene. 