|
||||||
|
 | |
 | |
 | |
 | Open reading frame (ORF) annotation in the exon skipping event |
 | |
 | 3'-UTR located exon skipping events lost miRNA binding sites |
 | |
 | |
 | Splicing Quantitative Trait Loci (sQTLs) in the skipped exons |
 | |
 | |
 |
Gene summary for MARK2 |
 Gene summary Gene summary |
| Gene information | Gene symbol | MARK2 | Gene ID | 2011 |
| Gene name | microtubule affinity regulating kinase 2 | |
| Synonyms | EMK-1|EMK1|PAR-1|Par-1b|Par1b | |
| Cytomap | 11q13.1 | |
| Type of gene | protein-coding | |
| Description | serine/threonine-protein kinase MARK2ELKL motif kinase 1MAP/microtubule affinity-regulating kinase 2PAR1 homolog bSer/Thr protein kinase PAR-1Bserine/threonine protein kinase EMKtesticular tissue protein Li 117 | |
| Modification date | 20200329 | |
| UniProtAcc | A0A024R555, | |
| Context | - 30930738(Genetic Overlap Between Alzheimer's Disease and Bipolar Disorder Implicates the MARK2 and VAC14 Genes) |
 Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Gene | GO ID | GO term | PubMed ID |
| MARK2 | GO:0006468 | protein phosphorylation | 14976552 |
| MARK2 | GO:0010976 | positive regulation of neuron projection development | 12429843 |
| MARK2 | GO:0018105 | peptidyl-serine phosphorylation | 10542369|16717194 |
| MARK2 | GO:0030010 | establishment of cell polarity | 12429843 |
| MARK2 | GO:0035556 | intracellular signal transduction | 14976552 |
| MARK2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 15324659 |
| MARK2 | GO:0070507 | regulation of microtubule cytoskeleton organization | 10542369 |
Top |
Gene structures and expression levels for MARK2 |
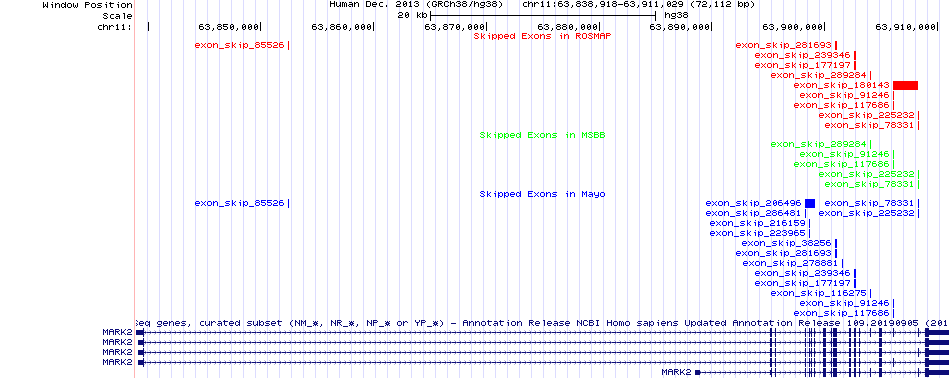
 Skipped exons in the ROSMAP, MSBB, and Mayo based on Ensembl gene isoform structure. Skipped exons in the ROSMAP, MSBB, and Mayo based on Ensembl gene isoform structure. * Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
ENSG00000072518
 Differentially expressed gene analysis across multiple brain tissues between AD and control. Differentially expressed gene analysis across multiple brain tissues between AD and control. |
| Tissue type | DEG direction | Base mean exp. | log2FC(AD/control) | P-value | Adj. p-value |
 Differentially expressed isoform analysis across multiple brain tissues between AD and control. Differentially expressed isoform analysis across multiple brain tissues between AD and control. |
| Tissue type | DEG direction | ENST | Transcript info. | Base mean exp. | log2FC(AD/control) | P-value | Adjc. p-value |
| CB | UP | ENST00000350490.11 | MARK2-201:protein_coding:MARK2 | 2.696761e+02 | 8.311876e-01 | 5.519004e-13 | 9.127737e-11 |
 Landscape of isoform expressions across multiple brain tissues between AD and control. Landscape of isoform expressions across multiple brain tissues between AD and control. |
Top |
Exon skipping events with PSIs in ROSMAP, MSBB, and Mayo for MARK2 |
 Landscape of individual exon skipping event across AD tissues and controls (PSI heatmap). Landscape of individual exon skipping event across AD tissues and controls (PSI heatmap). |
 All exon skipping events in AD cohorts. All exon skipping events in AD cohorts. |
| Exon skip ID | chr | Exons involved in exon skipping | Skipped exon |
| exon_skip_116275 | chr11 | 63903061:63903158:63903986:63904147:63904786:63904921 | 63903986:63904147 |
| exon_skip_117686 | chr11 | 63904786:63905043:63906088:63906114:63908260:63908304 | 63906088:63906114 |
| exon_skip_177197 | chr11 | 63902198:63902330:63902604:63902782:63903061:63903132 | 63902604:63902782 |
| exon_skip_216159 | chr11 | 63898232:63898280:63898608:63898673:63898763:63898793 | 63898608:63898673 |
| exon_skip_225232 | chr11 | 63906088:63906114:63908260:63908304:63908877:63909237 | 63908260:63908304 |
| exon_skip_239346 | chr11 | 63902198:63902330:63902601:63902782:63903061:63903132 | 63902601:63902782 |
| exon_skip_281693 | chr11 | 63900780:63900879:63900957:63901069:63902198:63902330 | 63900957:63901069 |
| exon_skip_286481 | chr11 | 63895580:63895633:63898232:63898280:63898608:63898673 | 63898232:63898280 |
| exon_skip_289284 | chr11 | 63903061:63903158:63903986:63904147:63904786:63905043 | 63903986:63904147 |
| exon_skip_78331 | chr11 | 63904786:63905043:63908260:63908304:63908877:63909237 | 63908260:63908304 |
| exon_skip_91246 | chr11 | 63904786:63905043:63906088:63906114:63908877:63909237 | 63906088:63906114 |
 Differentially expressed PSI values of individual exon skipping events in multiple brain tissues between AD and control. Differentially expressed PSI values of individual exon skipping events in multiple brain tissues between AD and control. |
| Exon skipping information | Tissue type | Avg(PSIs) in AD | Avg(PSIs) in control | Difference (PSI) | Adj. p-value |
| exon_skip_78331 | Mayo_CB | 3.352439e-01 | 4.923377e-01 | -1.570938e-01 | 1.197614e-05 |
Top |
Open reading frame (ORF) annotation in the exon skipping event for MARK2 |
 Open reading frame (ORF) of individual exon skipping events in ROSMAP based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in ROSMAP based on the Ensembl gene structure combined from isoforms. |
| ENST | Start of skipped exon | End of skipped exon | ORF |
| ENST00000402010 | 63900957 | 63901069 | Frame-shift |
| ENST00000402010 | 63902601 | 63902782 | Frame-shift |
| ENST00000402010 | 63903986 | 63904147 | In-frame |
| ENST00000402010 | 63906088 | 63906114 | In-frame |
| ENST00000402010 | 63908260 | 63908304 | In-frame |
 Open reading frame (ORF) of individual exon skipping events in MSBB based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in MSBB based on the Ensembl gene structure combined from isoforms. |
| ENST | Start of skipped exon | End of skipped exon | ORF |
| ENST00000402010 | 63903986 | 63904147 | In-frame |
| ENST00000402010 | 63906088 | 63906114 | In-frame |
| ENST00000402010 | 63908260 | 63908304 | In-frame |
 Open reading frame (ORF) of individual exon skipping events in Mayo based on the Ensembl gene structure combined from isoforms. Open reading frame (ORF) of individual exon skipping events in Mayo based on the Ensembl gene structure combined from isoforms. |
| ENST | Start of skipped exon | End of skipped exon | ORF |
| ENST00000402010 | 63898232 | 63898280 | Frame-shift |
| ENST00000402010 | 63900957 | 63901069 | Frame-shift |
| ENST00000402010 | 63902601 | 63902782 | Frame-shift |
| ENST00000402010 | 63898608 | 63898673 | In-frame |
| ENST00000402010 | 63903986 | 63904147 | In-frame |
| ENST00000402010 | 63906088 | 63906114 | In-frame |
| ENST00000402010 | 63908260 | 63908304 | In-frame |
Top |
Infer the effects of exon skipping event on protein functional features for MARK2 |
p-ENSG00000072518_img4.png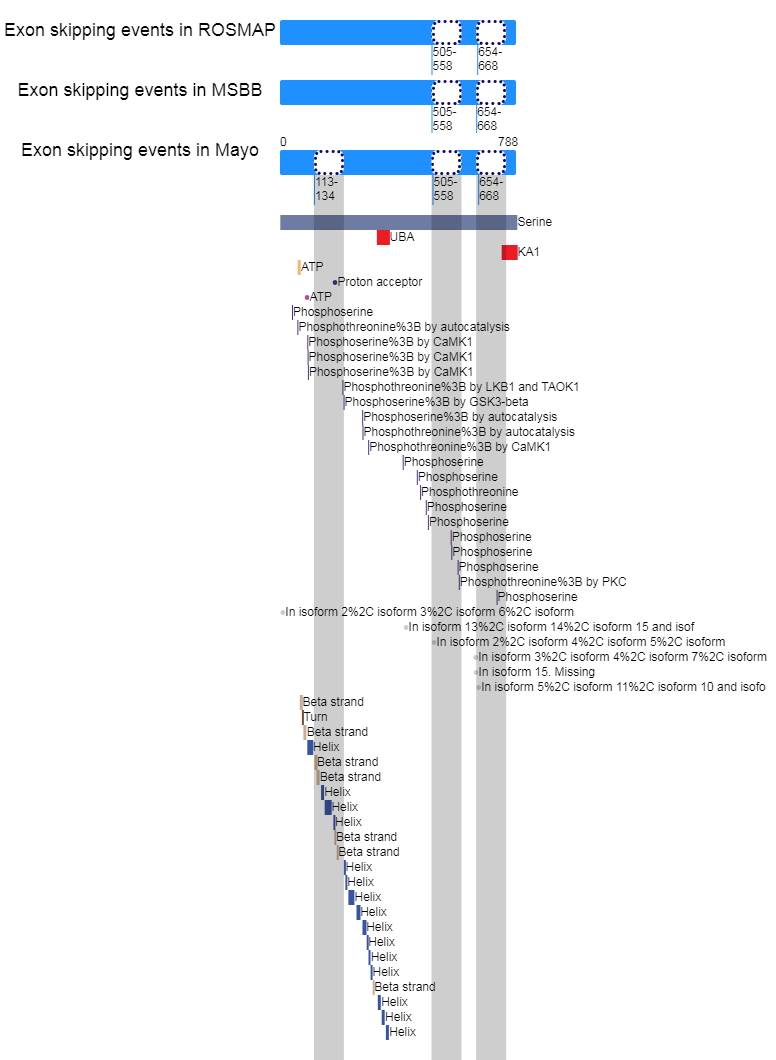 |
 Loci of skipped exons in ROSMAP across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in ROSMAP across genomic, transcript, and protein sequence levels of In-frame cases. |
| ENST | Length of mRNA | Length of AA seq. | Genomic start | Genomic end | mRNA start | mRNA end | AA start | AA end |
| ENST00000402010 | 4745 | 788 | 63903986 | 63904147 | 2095 | 2255 | 505 | 558 |
| ENST00000402010 | 4745 | 788 | 63908260 | 63908304 | 2542 | 2585 | 654 | 668 |
 Loci of skipped exons in MSBB across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in MSBB across genomic, transcript, and protein sequence levels of In-frame cases. |
| ENST | Length of mRNA | Length of AA seq. | Genomic start | Genomic end | mRNA start | mRNA end | AA start | AA end |
| ENST00000402010 | 4745 | 788 | 63903986 | 63904147 | 2095 | 2255 | 505 | 558 |
| ENST00000402010 | 4745 | 788 | 63908260 | 63908304 | 2542 | 2585 | 654 | 668 |
 Loci of skipped exons in Mayo across genomic, transcript, and protein sequence levels of In-frame cases. Loci of skipped exons in Mayo across genomic, transcript, and protein sequence levels of In-frame cases. |
| ENST | Length of mRNA | Length of AA seq. | Genomic start | Genomic end | mRNA start | mRNA end | AA start | AA end |
| ENST00000402010 | 4745 | 788 | 63898608 | 63898673 | 918 | 982 | 113 | 134 |
| ENST00000402010 | 4745 | 788 | 63903986 | 63904147 | 2095 | 2255 | 505 | 558 |
| ENST00000402010 | 4745 | 788 | 63908260 | 63908304 | 2542 | 2585 | 654 | 668 |
 Lost protein functional features of individual exon skipping events in ROSMAP. Lost protein functional features of individual exon skipping events in ROSMAP. |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q7KZI7 | 505 | 558 | 505 | 558 | Alternative sequence | ID=VSP_051706;Note=In isoform 2%2C isoform 4%2C isoform 5%2C isoform 7%2C isoform 9%2C isoform 10%2C isoform 13%2C isoform 15 and isoform 16. Missing;Ontology_term=ECO:0000303,ECO:0000303,ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:11433294,ECO:00 |
| Q7KZI7 | 505 | 558 | 1 | 788 | Chain | ID=PRO_0000086301;Note=Serine/threonine-protein kinase MARK2 |
| Q7KZI7 | 654 | 668 | 645 | 668 | Alternative sequence | ID=VSP_041853;Note=In isoform 15. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q7KZI7 | 654 | 668 | 654 | 668 | Alternative sequence | ID=VSP_051708;Note=In isoform 5%2C isoform 11%2C isoform 10 and isoform 12. Missing;Ontology_term=ECO:0000303,ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:11433294,ECO:0000303|PubMed:9730619,ECO:0000303|Ref.2;Dbxref=PMID:11433294,PMID:9730619 |
| Q7KZI7 | 654 | 668 | 1 | 788 | Chain | ID=PRO_0000086301;Note=Serine/threonine-protein kinase MARK2 |
 Lost protein functional features of individual exon skipping events in MSBB. Lost protein functional features of individual exon skipping events in MSBB. |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q7KZI7 | 505 | 558 | 505 | 558 | Alternative sequence | ID=VSP_051706;Note=In isoform 2%2C isoform 4%2C isoform 5%2C isoform 7%2C isoform 9%2C isoform 10%2C isoform 13%2C isoform 15 and isoform 16. Missing;Ontology_term=ECO:0000303,ECO:0000303,ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:11433294,ECO:00 |
| Q7KZI7 | 505 | 558 | 1 | 788 | Chain | ID=PRO_0000086301;Note=Serine/threonine-protein kinase MARK2 |
| Q7KZI7 | 654 | 668 | 645 | 668 | Alternative sequence | ID=VSP_041853;Note=In isoform 15. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q7KZI7 | 654 | 668 | 654 | 668 | Alternative sequence | ID=VSP_051708;Note=In isoform 5%2C isoform 11%2C isoform 10 and isoform 12. Missing;Ontology_term=ECO:0000303,ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:11433294,ECO:0000303|PubMed:9730619,ECO:0000303|Ref.2;Dbxref=PMID:11433294,PMID:9730619 |
| Q7KZI7 | 654 | 668 | 1 | 788 | Chain | ID=PRO_0000086301;Note=Serine/threonine-protein kinase MARK2 |
 Lost protein functional features of individual exon skipping events in Mayo. Lost protein functional features of individual exon skipping events in Mayo. |
| UniProt acc. | Start of exon skipping (AA) | End of exon skipping (AA) | Protein feature start (AA) | Protein feature end (AA) | Category of protein feature | Description of feature |
| Q7KZI7 | 113 | 134 | 115 | 120 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3IEC |
| Q7KZI7 | 113 | 134 | 122 | 129 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3IEC |
| Q7KZI7 | 113 | 134 | 1 | 788 | Chain | ID=PRO_0000086301;Note=Serine/threonine-protein kinase MARK2 |
| Q7KZI7 | 113 | 134 | 53 | 304 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Q7KZI7 | 505 | 558 | 505 | 558 | Alternative sequence | ID=VSP_051706;Note=In isoform 2%2C isoform 4%2C isoform 5%2C isoform 7%2C isoform 9%2C isoform 10%2C isoform 13%2C isoform 15 and isoform 16. Missing;Ontology_term=ECO:0000303,ECO:0000303,ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:11433294,ECO:00 |
| Q7KZI7 | 505 | 558 | 1 | 788 | Chain | ID=PRO_0000086301;Note=Serine/threonine-protein kinase MARK2 |
| Q7KZI7 | 654 | 668 | 645 | 668 | Alternative sequence | ID=VSP_041853;Note=In isoform 15. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| Q7KZI7 | 654 | 668 | 654 | 668 | Alternative sequence | ID=VSP_051708;Note=In isoform 5%2C isoform 11%2C isoform 10 and isoform 12. Missing;Ontology_term=ECO:0000303,ECO:0000303,ECO:0000303;evidence=ECO:0000303|PubMed:11433294,ECO:0000303|PubMed:9730619,ECO:0000303|Ref.2;Dbxref=PMID:11433294,PMID:9730619 |
| Q7KZI7 | 654 | 668 | 1 | 788 | Chain | ID=PRO_0000086301;Note=Serine/threonine-protein kinase MARK2 |
Top |
3'-UTR located exon skipping events that lost miRNA binding sites in MARK2 |
 3'-UTR exon skipping evnets lost miRNA binding. 3'-UTR exon skipping evnets lost miRNA binding. |
| Tissue type | ENST | Exon skip start | Exon skip end | microRNA | Binding site by TargetScan | Binding type by TargetScan | Bdinding site by miRanda | Score of miRanda | Energy by miRanda |
Top |
SNVs in the skipped exons for MARK2 |
 - Differential PSIs between mutated versus non-mutated samples. - Differential PSIs between mutated versus non-mutated samples. |
 - Depth of Coverage in the skipped exon of the mutated samples. - Depth of Coverage in the skipped exon of the mutated samples. |
 - Sashimi plot in the skipped exon of the mutated samples. - Sashimi plot in the skipped exon of the mutated samples. |
 - Non-synonymous mutations located in the skipped exons. - Non-synonymous mutations located in the skipped exons. |
| Cancer type | Sample | ESID | Skipped exon start | Skipped exon end | Mutation start | Mutation end | Mutation type | Reference seq | Mutation seq | AAchange |
 - Non-synonymous mutations located in the skipped exons in CCLE. - Non-synonymous mutations located in the skipped exons in CCLE. |
| Sample | Skipped exon start | Skipped exon end | Mutation start | Mutation end | Mutation type | Reference seq | Mutation seq | AAchange |
Top |
AD stage-associated exon skippint events for MARK2 |
 Associated exon skipping events with Braak staging or Clinical Dementia Rating (CDR). Associated exon skipping events with Braak staging or Clinical Dementia Rating (CDR). |
| AD stage info | Cohort | Tissue | SE id | Coefficient | P-value | Chromosome | Strand | E1 start | E1 end | Skipped start | Skipped end | E2 start | E2 end |
Top |
Splicing Quantitative Trait Loci (sQTL) in the exon skipping event for MARK2 |
 sQTL information located at the skipped exons. sQTL information located at the skipped exons. |
| Tissue type | Exon skip ID | SNP id | Location | P-value | FDR |
Top |
Correlation with RNA binding proteins (RBPs) for MARK2 |
 Correlated RBP and related information. Correlated RBP and related information. |
| Tissue type | RBP name | Exon skip ID | Correlation coeifficient | P-value |
| CB | CNOT4 | exon_skip_116275 | -4.336543e-01 | 1.129423e-08 |
| CB | PCBP4 | exon_skip_116275 | 4.648662e-01 | 6.698872e-10 |
| CB | TRA2A | exon_skip_91246 | -4.679452e-01 | 4.990538e-10 |
| CB | NUP42 | exon_skip_91246 | 4.631784e-01 | 7.862343e-10 |
| CB | TARDBP | exon_skip_78331 | -4.342362e-01 | 1.074237e-08 |
| CB | TRA2A | exon_skip_78331 | -6.177972e-01 | 4.156659e-18 |
| CB | NUP42 | exon_skip_78331 | 4.983905e-01 | 2.307250e-11 |
| HCC | SFPQ | exon_skip_289284 | -4.446892e-01 | 1.817010e-14 |
| HCC | RBM6 | exon_skip_289284 | -4.253032e-01 | 3.049649e-13 |
| HCC | EIF4G2 | exon_skip_289284 | -4.181902e-01 | 8.211360e-13 |
| IFG | PCBP4 | exon_skip_289284 | -4.520912e-01 | 1.572240e-02 |
| IFG | TARDBP | exon_skip_78331 | -5.846955e-01 | 1.360194e-03 |
| IFG | RBM5 | exon_skip_78331 | -5.190165e-01 | 5.536647e-03 |
| IFG | TRA2A | exon_skip_78331 | -4.957997e-01 | 8.539413e-03 |
| TC | NOVA1 | exon_skip_116275 | 4.721114e-01 | 2.931105e-10 |
Top |
RelatedDrugs for MARK2 |
 Approved drugs targeting this gene. Approved drugs targeting this gene. (DrugBank Version 5.1.0 2018-04-02) |
| UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Q7KZI7 | approved|investigational | DB12010 | Fostamatinib | small molecule | Q7KZI7 |
Top |
RelatedDiseases for MARK2 |
 Diseases associated with this gene. Diseases associated with this gene. (DisGeNet 4.0) |
| Gene | Disease ID | Disease name | # pubmeds | Source |
