|
Home |
Download |
Statistics |
Landscape |
Help |
Contact |
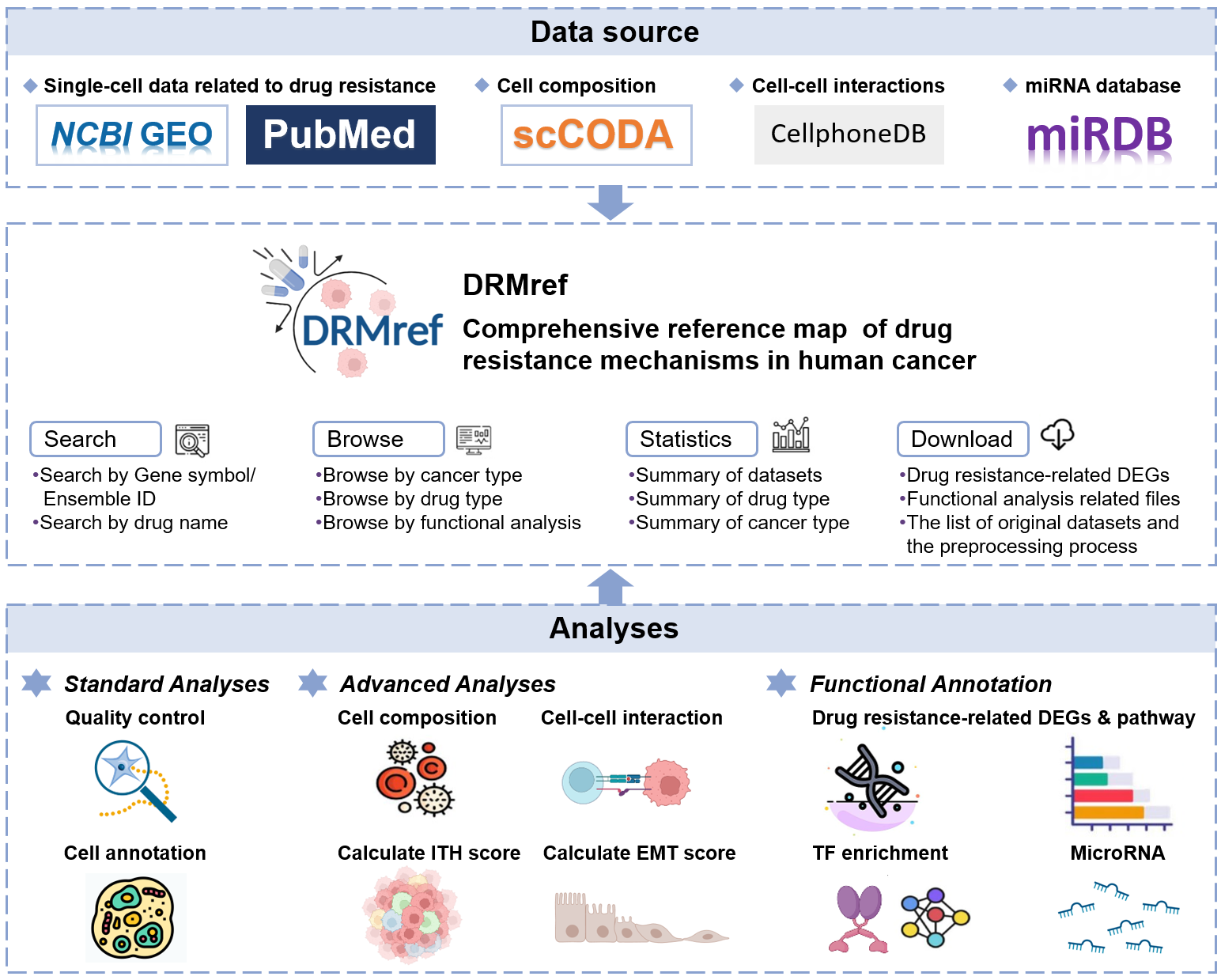
About DRMref
 Search
Search Browse by cancer type
Browse by cancer typeEach entry shows the datasets with this cancer type. The duplicate datasets represent that these datasets contain both pre- and post-treatment samples.
 Browse by drug type
Browse by drug typeEach entry shows the datasets with this drug type. The duplicate datasets represent that these datasets contain both pre- and post-treatment samples.
| • Chemotherapy (7) | • Targeted therapy (23) | • Immunotherapy (12) |
 Browse by functional analysis
Browse by functional analysisEach entry shows the unique datasets with this analysis. Additionally, the "Enrichment analysis of six known drug resistance mechanisms" module shows the differentially expressed genes (DEGs) enriched in six known drug resistance mechanisms. The "Difference of cell-cell interactions between resistant and sensitive groups" module also shows the DEGs involved in significant ligand-receptor pairs. The "Motifs and transcription factors(TFs) regulating drug resistance-related DEGs in each cell type" module also shows the differentially expressed TFs between the resistant and sensitive groups. "Differentially expressed drug target genes between the resistant and sensitive groups." module displays the DEGs that act as drug targets.