
|
||||||
|
 | |
 | |
 | Expression of this gene in the resistant group across all datasets and cell types |
 | Significant ligand-receptor pairs related to this gene (This gene does not contain this module) |
 | Known drug resistance mechanisms of this gene (This gene does not contain this module) |
 | |
 | |
 | Acts as a transcription factor (This gene does not contain this module) |
 | Acts as a drug target< (This gene does not contain this module) |
Gene: POLH |
Summary for POLH |
| Gene information | Gene symbol | POLH | Ensembl ID | ENSG00000170734 |
| Entrez ID | 5429 | |
| Gene name | DNA polymerase eta | |
| Synonyms | RAD30A|XP-V | |
| Gene type | protein_coding | |
| UniProtAcc | Q9Y253 |
Top |
Dataset with differentially expressed gene: POLH |
 Differentially expression genes between the resistant and sensitive groups. A positive avg_log2FC represents significantly high expression in the resistant group, while a negative avg_log2FC represents significantly low expression in the resistant group. Differentially expression genes between the resistant and sensitive groups. A positive avg_log2FC represents significantly high expression in the resistant group, while a negative avg_log2FC represents significantly low expression in the resistant group. |
| Dataset | Source | Tissue | Cancer type level1 | Cancer type level2 | Drug type | Regimen | Timepoint | Cell type | avg_log2FC | p_val_adj |
GSE169246_PacAteTissue | patients | Tumor tissue | Breast cancer | Advanced triple-negative breast cancer (TNBC) | Immunotherapy | atezolizumab + paclitaxel | post | Mast cells | -0.485427 | 4.62e-02 |
Top |
Expression of POLH in the resistant group across all datasets and cell types |
 Red dots represent significantly high expression in the resistant group, while blue dots represent significantly low expression in the resistant group. White dots represent that there is no significant difference in the expression of this gene between the resistant and sensitive groups. Blank represents that this cell type is not present in this dataset. (If the image exists, the user can click on it to enlarge it in a new window.) Red dots represent significantly high expression in the resistant group, while blue dots represent significantly low expression in the resistant group. White dots represent that there is no significant difference in the expression of this gene between the resistant and sensitive groups. Blank represents that this cell type is not present in this dataset. (If the image exists, the user can click on it to enlarge it in a new window.) |
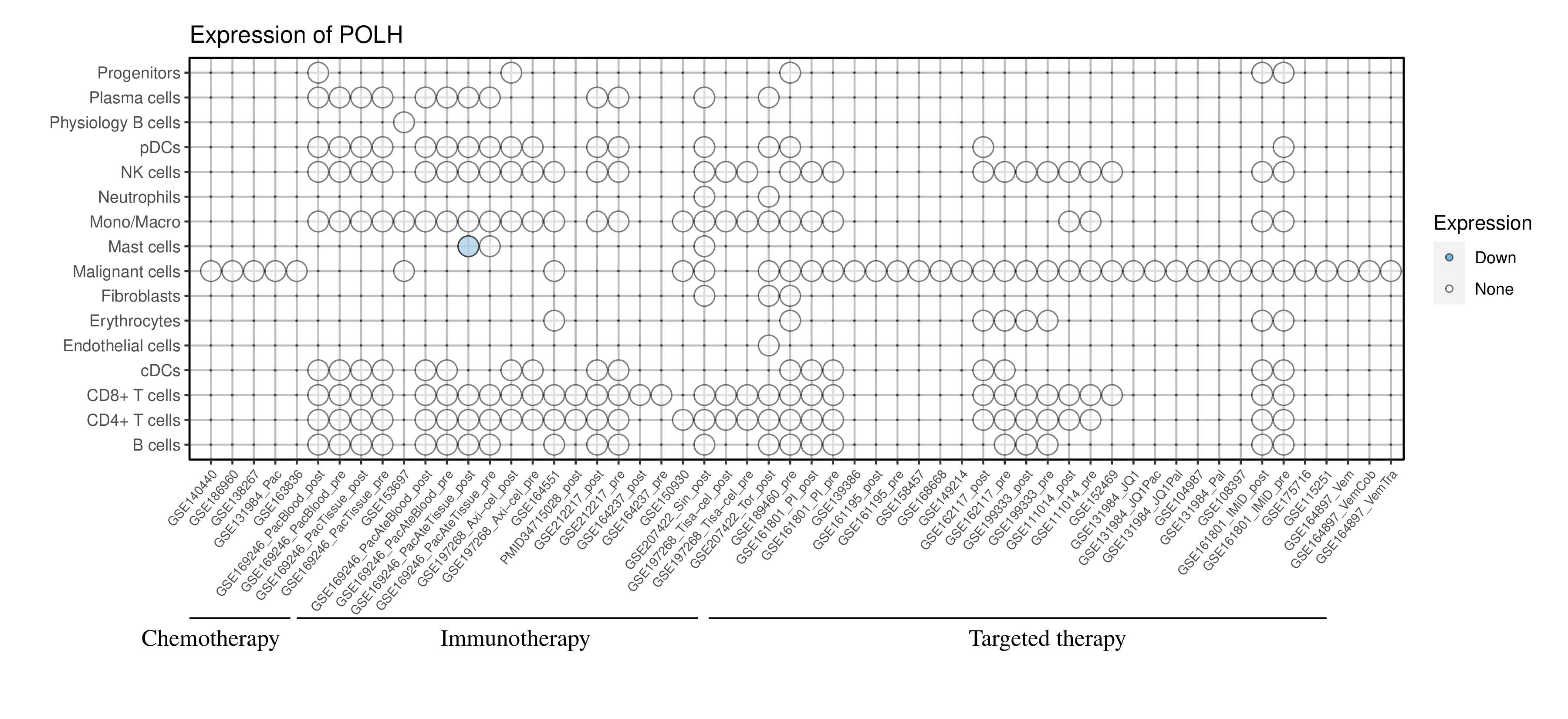 |
Top |
Significant ligand-receptor pairs related to POLH |
 This table shows the significant ligand-receptor pairs related to this gene across all datasets, timepoints and conditions. (Complete files can be downloaded from the download section.) This table shows the significant ligand-receptor pairs related to this gene across all datasets, timepoints and conditions. (Complete files can be downloaded from the download section.) |
Top |
Known drug resistance mechanisms of this gene |
 This table shows the known drug resistance mechanisms of this gene. Clicking on this gene will link to another database that displays more drug resistance-related information. This table shows the known drug resistance mechanisms of this gene. Clicking on this gene will link to another database that displays more drug resistance-related information. |
Top |
MicroRNAs (miRNA) regulating POLH |
 This table shows the miRNAs with a score of more than 80 regulating this gene. (Complete files can be downloaded from the download section.) This table shows the miRNAs with a score of more than 80 regulating this gene. (Complete files can be downloaded from the download section.) |
| Gene symbol | miRNA | Score | GeneBank accession |
| POLH | hsa-miR-6778-3p | 99.7391 | NM_001291969 |
| POLH | hsa-miR-6738-3p | 97.1738 | NM_001291970 |
| POLH | hsa-miR-577 | 96.5203 | NM_001291969 |
| POLH | hsa-miR-5001-3p | 96.455 | NM_001291970 |
| POLH | hsa-miR-616-5p | 95.9912 | NM_001291969 |
| POLH | hsa-miR-371b-5p | 95.9912 | NM_001291969 |
| POLH | hsa-miR-373-5p | 95.9912 | NM_001291969 |
| POLH | hsa-miR-3686 | 95.8909 | NM_001291969 |
| POLH | hsa-miR-4278 | 95.2845 | NM_001291969 |
| POLH | hsa-miR-1297 | 95.0844 | NM_001291969 |
| POLH | hsa-miR-12136 | 94.3674 | NM_001291969 |
| POLH | hsa-miR-4772-3p | 92.952 | NM_001291970 |
| POLH | hsa-miR-4728-5p | 92.9052 | NM_001291969 |
| POLH | hsa-miR-7151-3p | 92.782 | NM_001291969 |
| POLH | hsa-miR-619-5p | 92.5385 | NM_001291969 |
| POLH | hsa-miR-544a | 91.6214 | NM_001291970 |
| POLH | hsa-miR-6801-5p | 91.4035 | NM_001291969 |
| POLH | hsa-miR-10393-3p | 91.4035 | NM_001291969 |
| POLH | hsa-miR-6785-5p | 90.6512 | NM_001291969 |
| POLH | hsa-miR-6514-3p | 87.4984 | NM_001291969 |
| POLH | hsa-let-7a-2-3p | 87.152 | NM_001291969 |
| POLH | hsa-let-7g-3p | 87.152 | NM_001291969 |
| POLH | hsa-miR-26b-5p | 86.8438 | NM_001291969 |
| POLH | hsa-miR-26a-5p | 86.8438 | NM_001291969 |
| POLH | hsa-miR-4308 | 86.1972 | NM_001291970 |
| POLH | hsa-miR-149-3p | 86.0421 | NM_001291969 |
| POLH | hsa-miR-6883-5p | 86.0421 | NM_001291969 |
| POLH | hsa-miR-10522-5p | 84.475 | NM_001291970 |
| POLH | hsa-miR-504-3p | 83.3633 | NM_001291969 |
| POLH | hsa-miR-150-5p | 83.2325 | NM_001291969 |
| POLH | hsa-miR-548aw | 82.9246 | NM_001291970 |
| POLH | hsa-miR-4465 | 82.5929 | NM_001291969 |
| POLH | hsa-miR-3163 | 82.4124 | NM_001291969 |
| POLH | hsa-miR-10b-5p | 82.3267 | NM_001291969 |
| POLH | hsa-miR-10a-5p | 82.3267 | NM_001291969 |
| POLH | hsa-miR-6124 | 81.3022 | NM_001291969 |
| POLH | hsa-miR-6079 | 81.1891 | NM_001291970 |
| POLH | hsa-miR-9983-3p | 81.0286 | NM_001291969 |
| POLH | hsa-miR-6071 | 80.364 | NM_001291970 |
| POLH | hsa-miR-6811-3p | 80.1631 | NM_001291969 |
| POLH | hsa-miR-3662 | 80.0869 | NM_001291970 |
| Page: 1 |
Top |
Motifs and transcription factors (TFs) regulating POLH |
 This table shows the Motifs and transcription factors (TFs) regulating this gene. (Complete files can be downloaded from the download section.) This table shows the Motifs and transcription factors (TFs) regulating this gene. (Complete files can be downloaded from the download section.) |
| Gene symbol | motif | TF_highConf |
| POLH | transfac_pro__M05523 | ZBTB46 (directAnnotation). |
| POLH | swissregulon__hs__MYB | MYB (directAnnotation). |
| POLH | taipale_tf_pairs__ETV2_RFX5_NACTTCCGGYNNNNGCAACSN_CAP_repr | ETV2; RFX5 (directAnnotation). |
| POLH | transfac_pro__M04944 | GABPA (directAnnotation). |
| POLH | nitta__sens_TATGCT40NACT_KAA_AATAAATCACTGCA_m1_c3 | GFI1 (inferredBy_Orthology). |
| POLH | transfac_pro__M06340 | ZNF790 (directAnnotation). |
| POLH | tfdimers__MD00388 | LEF1; NKX3-2; TCF7; TCF7L1 (directAnnotation). |
| Page: 1 |
Top |
Acts as a transcription factor |
 This table shows that this differential gene acts as a transcription factor. This table shows that this differential gene acts as a transcription factor. |
Top |
Acts as a drug target |
 This table shows that this differential gene acts as a drug target. This table shows that this differential gene acts as a drug target. |
| 1 | "Sun X, Zhang Y, Li H, Zhou Y, Shi S, Chen Z, He X, Zhang H, Li F, Yin J, Mou M, Wang Y, Qiu Y, Zhu F. DRESIS: the first comprehensive landscape of drug resistance information. Nucleic Acids Res. 2023 Jan 6;51(D1):D1263-D1275. doi: 10.1093/nar/gkac812. PMID: 36243960; PMCID: PMC9825618." |
| 2 | "Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018 Jan 4;46(D1):D1074-D1082. doi: 10.1093/nar/gkx1037. PMID: 29126136; PMCID: PMC5753335." |
