 lncRNA targets the enhancer region and positively regulates the gene expression. lncRNA targets the enhancer region and positively regulates the gene expression. |
 lncRNA targets the promoter region and positively regulates the gene expression. lncRNA targets the promoter region and positively regulates the gene expression. |
 lncRNA targets the promoter region and negatively regulates the gene expression. lncRNA targets the promoter region and negatively regulates the gene expression. |
 lncRNA targets the 3'UTR region and negatively regulates the mRNA. lncRNA targets the 3'UTR region and negatively regulates the mRNA. |
| Only predicted by lncTar | | CNOT1,CLIP4,OCIAD1,DSTYK,TNS3,PBLD,CPQ,MBP,GSK3B,VPS41,DLST,VPS26C,APPL2,AP5M1,VPS4B,UHRF1BP1L,ADCY7,RNF123,SLC11A2,CHP1,HACD3,TBC1D5,NT5C2,ZMYND11,RALGAPB,MAP4K3,YPEL2,TOM1L2,BECN1,VWA5A,HIPK1,DPH1,RNF40,CDKL5,SECISBP2L,SCP2,UBR1,DAG1,MINDY1,NOP9,PCSK6,GNL3L,ANKRD46,SPIRE1,TAOK3,TMEM161B,ACVR1B,KIAA1109,MPPE1,VTI1B,GORASP1,SCYL2,TBC1D15,MAP9,PLEKHB2,ITPR2,ARHGAP44,FRG1BP,ARHGAP21,FCHO2,CREBBP,SCOC,UBE4A,PEX1,ACSL1,TRAK1,ABLIM1,ZSWIM8,PHACTR2,MAN2A2,VPS13B,NR2C2,LDB1,TK2,BTBD9,SEC24B,CLTC,ATRN,AP1AR,SACM1L,MAN1A2,PDHX,SDHC,ALG12,FAM91A1,VGLL4,SON,KMT2D,PIK3C3,THTPA,NBR1,BCL2L13,TMPRSS2,SNX19,PKD2,PDPK1,EXOC7,MCCC2,AP3D1,ARFIP1,MON2,USP33,ACBD5,PMS2,AFDN,CPEB4,LONP2,TP53BP1,PCNX1,ERLIN2,PTPN4,PPIP5K2,TTC3,PC,LRP10,CNNM2,LYRM7,KIDINS220,DYNC1I2,ABRAXAS1,AKTIP,RASSF8,TMCO3,TTBK2,KIF13A,RNF31,CLIP1,CMTR2,CLMN,RFFL,RAD50,ABAT,CPT1A,ETFDH,PCCA,IQGAP2,RAI1,KIAA0319L,NCOA2,DCAF11,MKLN1,ARHGEF9,DYNC2H1,ARHGAP5,CLCC1,NKIRAS1,BCL2L2,MPDZ,CCSER2,ITSN2,ASXL2,TMEM220,THAP6,NBPF3,RNF169,MTMR12,CCDC85C,ALAS1,KLF13,WDR81,POLI,CRY2,TM9SF1,MFAP3L,UTRN,BAZ2A,USP9X,MYOF,ZFYVE1,KIF13B,UEVLD,DYNC1LI2,SLC35A5,KBTBD4,MIDEAS,CCDC121,ZNF106,MTO1,RASA1,APP,DENND4A,ARL1,IQSEC1,RREB1,MED13,CERT1,VPS50,MTARC2,WDR7,REEP5,SMG6,SF3A1,PHC3,KIAA0232,TMLHE,MYO10,DDB1,SIK3,FICD,NAAA,EPS15,AZI2,TIRAP,DOCK9,SYNJ1,PLSCR4,MGAT4A,PPARA,GFM2,PCMTD1,ATP9B,GPATCH8,VAMP7,PTPRM,GALC,PIGN,GTF2A1,ALDH5A1,TFIP11,PRKAA1,WDR72,OGDH,EIF3L,HBP1,SOS2,CSNK1A1,YTHDF3,LARS2,FAM160B1,MAPK9,BACE1,ATP1A1,HEATR5B,POLK,PRKAR1A,ACADM,PPM1A,TTC37,F11,FAXDC2,ETFRF1,DENND5B,ELF1,IL6ST,MIEF1,CITED2,MECP2,BLOC1S6,AFF1,TAF2,ARL2BP,GCC2,DCAF5,CHD6,USP8,TEX2,LMO7,BCOR,BDP1,RAD17,DPY19L3,FAM114A2,FAM13B,SNX18,TCEAL1,ITFG1,BTD,HEATR5A,C6orf89,RUSF1,GABARAPL1,TMBIM6,ATF1,ARHGAP32,ILDR1,KIAA1191,FAM172A,BCKDHB,DOCK1,RPRD1A,ALDH2,ZNF91,MAN2B2,MCFD2,RALGAPA1,FER,IBTK,NDUFS1,FNBP1L,SLC39A6,SCAF11,CDH1,RAB6A | | Exist in public source | | NA |
 lncRNA targets the skipped exon region. lncRNA targets the skipped exon region. | | -lncRNA and exonskipping events are positively correlated. |
| LncRNA Ensembl ID | LncRNA ENST ID | Exon ID | Skipped Exon | dG | ndG | Gene name with skipped exon | TransID with skipped exon | ORF mutation | | ENSG00000280206 | ENST00000624682 | exon_skip_361434 | chr21:25997359-25997416 | -8.08 | -0.1757 | APP | ENST00000346798 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_470719 | chr7:117134123-117134192 | -9.17 | -0.1798 | ST7 | ENST00000265437 | In-frame |
| -lncRNA and exonskipping events are negatively correlated. |
 |
| LncRNA Ensembl ID | LncRNA ENST ID | Exon ID | Skipped Exon | dG | ndG | Gene name with skipped exon | TransID with skipped exon | LOF | | ENSG00000280206 | ENST00000624682 | exon_skip_4279 | chr1:36174897-36175008 | -12.68 | -0.1294 | MAP7D1 | ENST00000373151 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_27178 | chr1:53233541-53233652 | -11.47 | -0.1062 | MAGOH | ENST00000371470 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_48716 | chr10:34372497-34372536 | -7.38 | -0.1995 | PARD3 | ENST00000374789 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_76344 | chr11:83278725-83278829 | -9.75 | -0.1026 | CCDC90B | ENST00000529689 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_83404 | chr12:53465931-53465973 | -10.15 | -0.3904 | PCBP2 | ENST00000439930 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_111672 | chr14:23089981-23090101 | -18.67 | -0.2122 | ACIN1 | ENST00000262710 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_153096 | chr17:44074968-44075087 | -15.63 | -0.1579 | G6PC3 | ENST00000269097 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_303345 | chr19:17227839-17228005 | -16.21 | -0.1059 | OCEL1 | ENST00000215061 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_319500 | chr19:45413962-45414034 | -9.73 | -0.1622 | ERCC1 | ENST00000300853,ENST00000589165 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_320509 | chr19:48839204-48839263 | -9.16 | -0.2411 | PLEKHA4 | ENST00000263265 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_430636 | chr4:82862533-82862572 | -7.50 | -0.9375 | SEC31A | ENST00000355196,ENST00000395310,ENST00000448323 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_462160 | chr6:127316449-127316502 | -6.80 | -0.6182 | ECHDC1 | ENST00000531967 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_5818 | chr1:45568484-45568684 | -19.20 | -0.1067 | AKR1A1 | ENST00000351829,ENST00000372070 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_85538 | chr12:71894324-71894375 | -8.52 | -0.3408 | TBC1D15 | ENST00000550746 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_110809 | chr14:105449358-105449409 | -10.67 | -0.3334 | MTA1 | ENST00000331320 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_382458 | chr3:38131595-38131638 | -6.61 | -0.1612 | ACAA1 | ENST00000333167 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_2091 | chr1:17634547-17634562 | -5.21 | -1.0420 | ARHGEF10L | ENST00000361221 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_11029 | chr1:154989559-154989707 | -20.78 | -0.1676 | FLAD1 | ENST00000292180 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_27065 | chr1:52906518-52906611 | -13.18 | -0.2197 | ECHDC2 | ENST00000371522 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_30682 | chr1:151310210-151310255 | -9.67 | -0.2249 | PI4KB | ENST00000368873 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_33006 | chr1:156294758-156295016 | -24.13 | -0.1001 | GLMP | ENST00000362007 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_67681 | chr11:893426-893519 | -16.25 | -0.1766 | CHID1 | ENST00000323578,ENST00000436108,ENST00000449825 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_87733 | chr12:120571190-120571291 | -11.97 | -0.1962 | RNF10 | ENST00000325954 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_89597 | chr12:6672349-6672532 | -21.19 | -0.1292 | ZNF384 | ENST00000361959,ENST00000396801 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_140339 | chr16:635611-635774 | -21.48 | -0.1579 | METTL26 | ENST00000301686 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_145647 | chr16:69121052-69121170 | -13.92 | -0.1180 | CHTF8 | ENST00000398235,ENST00000448552 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_292904 | chr17:69251746-69251866 | -11.28 | -0.1106 | ABCA5 | ENST00000392676,ENST00000588877 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_301267 | chr19:6746028-6746196 | -23.83 | -0.1690 | TRIP10 | ENST00000313244 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_331103 | chr2:171712766-171712826 | -7.87 | -0.1640 | DYNC1I2 | ENST00000397119,ENST00000409773 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_351126 | chr20:36214356-36214440 | -11.56 | -0.1752 | EPB41L1 | ENST00000338074 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_353131 | chr20:58669290-58669453 | -19.75 | -0.1344 | STX16 | ENST00000371141 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_430112 | chr4:75653982-75654081 | -13.41 | -0.1541 | G3BP2 | ENST00000359707,ENST00000395719 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_446770 | chr5:178215275-178215430 | -16.33 | -0.1081 | PHYKPL | ENST00000308158 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_75223 | chr11:72012400-72012442 | -8.30 | -0.2184 | NUMA1 | ENST00000393695 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_124988 | chr15:90890981-90891191 | -20.74 | -0.1103 | FES | ENST00000328850 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_423688 | chr4:53414614-53414722 | -9.24 | -0.1216 | FIP1L1 | ENST00000337488 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_423795 | chr4:55888887-55888932 | -6.60 | -0.1886 | EXOC1 | ENST00000346134,ENST00000381295 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_13857 | chr1:168038370-168038488 | -10.03 | -0.1320 | DCAF6 | ENST00000312263 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_453157 | chr6:75640683-75640704 | -5.54 | -0.4262 | SENP6 | ENST00000447266 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_26372 | chr1:46560233-46560277 | -8.90 | -0.2871 | MKNK1 | ENST00000371946 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_52891 | chr10:98429790-98429889 | -15.34 | -0.1763 | HPS1 | ENST00000325103,ENST00000361490 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_95500 | chr12:96003837-96003920 | -9.89 | -0.1393 | LTA4H | ENST00000228740 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_141626 | chr16:4821546-4821597 | -8.35 | -0.2530 | GLYR1 | ENST00000321919 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_153767 | chr17:48057031-48057121 | -18.87 | -0.2621 | NFE2L1 | ENST00000362042 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_339935 | chr2:70991829-70991983 | -20.33 | -0.1564 | TEX261 | ENST00000272438 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_390418 | chr3:183842297-183842447 | -12.82 | -0.1002 | PARL | ENST00000317096 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_507092 | chr9:127732563-127732691 | -15.43 | -0.1484 | TOR2A | ENST00000373284 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_509230 | chrX:13763744-13763855 | -12.21 | -0.1131 | OFD1 | ENST00000340096 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_6457 | chr1:52083339-52083541 | -12.81 | -0.1334 | BTF3L4 | ENST00000313334 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_11532 | chr1:155137560-155137722 | -16.45 | -0.1337 | SLC50A1 | ENST00000368404 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_31972 | chr1:154213892-154213946 | -7.14 | -0.2644 | C1orf43 | ENST00000368521 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_83512 | chr12:53505134-53505262 | -16.00 | -0.1524 | TARBP2 | ENST00000266987 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_91507 | chr12:47795905-47796016 | -15.01 | -0.1614 | HDAC7 | ENST00000427332 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_294727 | chr17:82019640-82019694 | -8.80 | -0.1913 | CENPX | ENST00000392359 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_299306 | chr18:50274185-50274353 | -19.05 | -0.1155 | MBD1 | ENST00000269468,ENST00000382948,ENST00000591416 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_322579 | chr19:55455635-55455845 | -21.83 | -0.1137 | ISOC2 | ENST00000425675 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_334374 | chr2:227347225-227347384 | -14.73 | -0.1052 | MFF | ENST00000353339 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_387819 | chr3:127575748-127575809 | -12.05 | -0.2274 | TPRA1 | ENST00000355552,ENST00000450633,ENST00000489960 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_461111 | chr6:90544551-90544632 | -9.68 | -0.1195 | MAP3K7 | ENST00000369329 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_482403 | chr8:27445793-27445919 | -14.25 | -0.1357 | PTK2B | ENST00000346049,ENST00000397501 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_484062 | chr8:73030335-73030395 | -8.59 | -0.2526 | TERF1 | ENST00000276603 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_489377 | chr8:38429681-38429948 | -31.63 | -0.1185 | FGFR1 | ENST00000447712 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_140283 | chr16:635611-635774 | -21.48 | -0.1579 | METTL26 | ENST00000301686 | Frame-shift | | ENSG00000280206 | ENST00000624682 | exon_skip_61261 | chr11:65713347-65713503 | -15.22 | -0.1119 | KAT5 | ENST00000377046 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_447940 | chr6:10751132-10751234 | -13.56 | -0.1370 | TMEM14B | ENST00000379542 | In-frame | | ENSG00000280206 | ENST00000624682 | exon_skip_508362 | chr9:136676584-136676680 | -14.09 | -0.1515 | AGPAT2 | ENST00000371696,ENST00000538402 | In-frame |
 lncRNA targets by miRNA. lncRNA targets by miRNA. |
 |
| LncRNA Ensembl ID | miRNA ID | LncRNA ENST ID | Binding site in lncRNA | Score | Energy | Align Len | Public source | | ENSG00000280206 | hsa-mir-1468 | ENST00000624682 | chr16:15701768-15701864 | 184.00 | -64.99 | 92 | NA | | ENSG00000280206 | hsa-mir-1468 | ENST00000624682 | chr16:15701315-15701400 | 172.00 | -62.88 | 84 | NA | | ENSG00000280206 | hsa-mir-1468 | ENST00000624682 | chr16:15701547-15701632 | 162.00 | -65.62 | 75 | NA | | ENSG00000280206 | hsa-mir-1468 | ENST00000624682 | chr16:15701457-15701543 | 152.00 | -69.86 | 87 | NA | | ENSG00000280206 | hsa-mir-1468 | ENST00000624682 | chr16:15702028-15702118 | 151.00 | -66.40 | 84 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15701483-15701574 | 200.00 | -62.65 | 92 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15701811-15701893 | 195.00 | -60.54 | 85 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15701697-15701778 | 192.00 | -58.15 | 84 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15701276-15701367 | 150.00 | -64.89 | 89 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15701581-15701679 | 150.00 | -73.59 | 91 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15701389-15701484 | 147.00 | -83.32 | 89 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15701930-15702017 | 145.00 | -63.03 | 90 | NA | | ENSG00000280206 | hsa-mir-29c | ENST00000624682 | chr16:15702011-15702101 | 145.00 | -62.60 | 87 | NA | | ENSG00000280206 | hsa-mir-139 | ENST00000624682 | chr16:15701618-15701684 | 167.00 | -58.22 | 64 | NA | | ENSG00000280206 | hsa-mir-139 | ENST00000624682 | chr16:15701262-15701338 | 159.00 | -65.16 | 70 | NA | | ENSG00000280206 | hsa-mir-139 | ENST00000624682 | chr16:15701414-15701484 | 155.00 | -69.37 | 59 | NA | | ENSG00000280206 | hsa-mir-139 | ENST00000624682 | chr16:15701695-15701776 | 147.00 | -53.91 | 79 | NA | | ENSG00000280206 | hsa-mir-22 | ENST00000624682 | chr16:15701673-15701755 | 164.00 | -68.90 | 84 | NA | | ENSG00000280206 | hsa-mir-22 | ENST00000624682 | chr16:15701238-15701293 | 149.00 | -56.68 | 40 | NA | | ENSG00000280206 | hsa-mir-22 | ENST00000624682 | chr16:15702025-15702119 | 145.00 | -74.14 | 84 | NA | | ENSG00000280206 | hsa-mir-22 | ENST00000624682 | chr16:15701307-15701397 | 144.00 | -72.86 | 88 | NA |
 RNA A-to-I editing events in lncRNA. RNA A-to-I editing events in lncRNA. |
| LncRNAediting ID | LncRNA Ensembl ID | Chromosome | Editing Position | Strand | Gene Type | Gene Name | Transcript ID | Transcript Type | Transcript Name |
 Edited-associated DElncRNAs in cancer. Edited-associated DElncRNAs in cancer. |
| LncRNA Ensembl ID | LncRNA Index | Cancer Type | Chr_Postion_Strand | AVE1 | AVE2 | log2FC | W-value | P-value | Adjc.p-value | Change |
 Correlation between RNA A-to-I editing events's frequecy and lncRNA expression. Correlation between RNA A-to-I editing events's frequecy and lncRNA expression. |
| LncRNA Ensembl ID | LncRNA Index | Correlation | P-value | Adjc.p-value |
 Cis-expression quantitative trait loci(cis-eQTL) of lncRNA. Cis-expression quantitative trait loci(cis-eQTL) of lncRNA. |
| LncRNA Ensembl ID | LncRNA Name | SNP info | Number of Positive corelated Cancer | Positive corelated Cancer | Number of Negative corelated Cancer | Negative corelated Cancer |
 lncRNA regulates differentially expressed genes by function as enhancer. lncRNA regulates differentially expressed genes by function as enhancer. |
| LncRNA Ensembl ID | PC Gene ID | PC Gene Name | Positive correlated cancers | Cancer with PC gene up-regulation | Cancer with PC gene down-regulation |
 LncRNA-TF complex positively regulates the gene expression by target promoter region (#cancer types with positive correlation >= 5). LncRNA-TF complex positively regulates the gene expression by target promoter region (#cancer types with positive correlation >= 5). |
| lncRNA ID | TF ID | TF Name | PCgene ID | PCgene Name | Number of Cancer | Canaer Types |
 LncRNA-TF complex negatively regulates the gene expression by target promoter region (#cancer types with negative correlation >= 5). LncRNA-TF complex negatively regulates the gene expression by target promoter region (#cancer types with negative correlation >= 5). |
| lncRNA ID | TF ID | TF Name | PCgene ID | PCgene Name | Number of Cancer | Canaer Types |
 LncRNA-RBP complex positively regulates the exon skippping events by target skipped eoxon region. LncRNA-RBP complex positively regulates the exon skippping events by target skipped eoxon region. |
| LncRNA Ensembl ID | RBP ID | RBP Gene Name | Exon Skipping ID | Skipped Exon | EX Gene Name | EX Affected TransID | ORF_anno | Cancer Type |
 LncRNA-RBP complex negatively regulates the exon skippping events by target skipped eoxon region. LncRNA-RBP complex negatively regulates the exon skippping events by target skipped eoxon region. |
| LncRNA Ensembl ID | RBP ID | RBP Gene Name | Exon Skipping ID | Skipped Exon | EX Gene Name | EX Affected TransID | ORF_anno | Cancer Type |
 lncRNA regulates differential expressed mRNA by directly targeting 3' UTR region. lncRNA regulates differential expressed mRNA by directly targeting 3' UTR region. |
| LncRNA Ensembl ID | LncRNA ENST ID | PC Gene Name | PC Gene ID | PC ENST ID | dG | nDG | Cancer with PC gene up-regulation | Cancer with PC gene Down-regulation | | ENSG00000280206 | ENST00000624682 | VWA5A | ENSG00000110002 | ENST00000392744 | -0.3424 | 359 | - | KIRC,HNSC,LUSC,COAD | | ENSG00000280206 | ENST00000624682 | ALDH2 | ENSG00000111275 | ENST00000416293 | -0.4476 | 518 | - | GBM,CHOL,HNSC,LUSC,LUAD,LIHC | | ENSG00000280206 | ENST00000624682 | MCCC2 | ENSG00000131844 | ENST00000509358 | -0.5377 | 336 | - | CHOL | | ENSG00000280206 | ENST00000624682 | APH1B | ENSG00000138613 | ENST00000380343 | -0.5842 | 169 | - | LUSC | | ENSG00000280206 | ENST00000624682 | IQSEC1 | ENSG00000144711 | ENST00000613206 | -0.1811 | 164 | - | GBM,CHOL,LUSC | | ENSG00000280206 | ENST00000624682 | FAXDC2 | ENSG00000170271 | ENST00000517938 | -0.1396 | 191 | - | GBM,CHOL,HNSC,ESCA,LUSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | FAXDC2 | ENSG00000170271 | ENST00000518651 | -0.2412 | 96 | - | GBM,CHOL,HNSC,ESCA,LUSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | FAXDC2 | ENSG00000170271 | ENST00000520968 | -0.7771 | 397 | - | GBM,CHOL,HNSC,ESCA,LUSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | ETFDH | ENSG00000171503 | ENST00000307738 | -0.1366 | 51 | - | CHOL,HNSC,ESCA,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | NT5C2 | ENSG00000076685 | ENST00000458345 | -0.1962 | 145 | - | HNSC | | ENSG00000280206 | ENST00000624682 | RNF123 | ENSG00000164068 | ENST00000432042 | -0.1436 | 184 | - | GBM,CHOL,HNSC | | ENSG00000280206 | ENST00000624682 | YPEL2 | ENSG00000175155 | ENST00000585166 | -0.1123 | 167 | - | GBM,LIHC | | ENSG00000280206 | ENST00000624682 | KIF13B | ENSG00000197892 | ENST00000523130 | -0.6571 | 580 | - | GBM,KIRC,HNSC,LUSC,COAD | | ENSG00000280206 | ENST00000624682 | SACM1L | ENSG00000211456 | ENST00000418611 | -0.1197 | 483 | - | CHOL,LUSC | | ENSG00000280206 | ENST00000624682 | NDUFS1 | ENSG00000023228 | ENST00000449699 | -0.1935 | 345 | - | CHOL,KIRC,HNSC | | ENSG00000280206 | ENST00000624682 | NDUFS1 | ENSG00000023228 | ENST00000432169 | -0.1935 | 345 | - | CHOL,KIRC,HNSC | | ENSG00000280206 | ENST00000624682 | ALAS1 | ENSG00000023330 | ENST00000493402 | -0.5664 | 162 | - | CHOL,KIRC,LIHC | | ENSG00000280206 | ENST00000624682 | BCKDHB | ENSG00000083123 | ENST00000369760 | -1.0680 | 120 | - | CHOL,KIRC,HNSC,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | F11 | ENSG00000088926 | ENST00000452239 | -0.1352 | 453 | - | CHOL,KIRC,KIRP,LIHC | | ENSG00000280206 | ENST00000624682 | DCAF11 | ENSG00000100897 | ENST00000560457 | -0.1247 | 131 | - | CHOL,KIRC | | ENSG00000280206 | ENST00000624682 | DCAF11 | ENSG00000100897 | ENST00000558706 | -0.1407 | 115 | - | CHOL,KIRC | | ENSG00000280206 | ENST00000624682 | DCAF11 | ENSG00000100897 | ENST00000560614 | -0.1016 | 72 | - | CHOL,KIRC | | ENSG00000280206 | ENST00000624682 | DCAF11 | ENSG00000100897 | ENST00000557888 | -0.1086 | 157 | - | CHOL,KIRC | | ENSG00000280206 | ENST00000624682 | LONP2 | ENSG00000102910 | ENST00000535754 | -0.1205 | 1 | - | CHOL | | ENSG00000280206 | ENST00000624682 | LONP2 | ENSG00000102910 | ENST00000416006 | -0.1088 | 414 | - | CHOL | | ENSG00000280206 | ENST00000624682 | MPDZ | ENSG00000107186 | ENST00000536827 | -0.1257 | 101 | - | CHOL,ESCA,LUSC,LIHC | | ENSG00000280206 | ENST00000624682 | MPDZ | ENSG00000107186 | ENST00000447879 | -0.1189 | 102 | - | CHOL,ESCA,LUSC,LIHC | | ENSG00000280206 | ENST00000624682 | MPDZ | ENSG00000107186 | ENST00000546205 | -0.4185 | 231 | - | CHOL,ESCA,LUSC,LIHC | | ENSG00000280206 | ENST00000624682 | PBLD | ENSG00000108187 | ENST00000468798 | -0.1141 | 101 | - | CHOL,KIRC,KIRP,ESCA,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | ALDH5A1 | ENSG00000112294 | ENST00000491546 | -0.1432 | 628 | - | GBM,CHOL,KIRC | | ENSG00000280206 | ENST00000624682 | PLSCR4 | ENSG00000114698 | ENST00000446574 | -0.1087 | 42 | - | CHOL,LUSC,LUAD,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | PLSCR4 | ENSG00000114698 | ENST00000493382 | -0.2931 | 86 | - | CHOL,LUSC,LUAD,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | SCP2 | ENSG00000116171 | ENST00000408941 | -0.1035 | 1 | - | CHOL,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | SCP2 | ENSG00000116171 | ENST00000484100 | -0.1035 | 1 | - | CHOL,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | ACADM | ENSG00000117054 | ENST00000420607 | -0.1235 | 502 | - | CHOL,KIRC,HNSC,KIRP,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | MTARC2 | ENSG00000117791 | ENST00000425560 | -0.1505 | 200 | - | CHOL,HNSC,KIRP,ESCA,LUSC,LUAD,COAD | | ENSG00000280206 | ENST00000624682 | ITPR2 | ENSG00000123104 | ENST00000242737 | -0.2916 | 737 | - | CHOL,KIRC,HNSC,KIRP | | ENSG00000280206 | ENST00000624682 | GABARAPL1 | ENSG00000139112 | ENST00000539408 | -0.1107 | 171 | - | GBM,CHOL,KIRC,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | GABARAPL1 | ENSG00000139112 | ENST00000544284 | -0.2068 | 117 | - | GBM,CHOL,KIRC,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | GABARAPL1 | ENSG00000139112 | ENST00000541453 | -0.1443 | 57 | - | GBM,CHOL,KIRC,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | GABARAPL1 | ENSG00000139112 | ENST00000543602 | -0.4925 | 154 | - | GBM,CHOL,KIRC,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | GABARAPL1 | ENSG00000139112 | ENST00000545887 | -0.1648 | 98 | - | GBM,CHOL,KIRC,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | TMBIM6 | ENSG00000139644 | ENST00000552370 | -0.3406 | 176 | - | CHOL | | ENSG00000280206 | ENST00000624682 | PCSK6 | ENSG00000140479 | ENST00000622483 | -0.1479 | 85 | - | CHOL,COAD | | ENSG00000280206 | ENST00000624682 | PCSK6 | ENSG00000140479 | ENST00000619160 | -0.1479 | 85 | - | CHOL,COAD | | ENSG00000280206 | ENST00000624682 | PCSK6 | ENSG00000140479 | ENST00000621185 | -0.8317 | 456 | - | CHOL,COAD | | ENSG00000280206 | ENST00000624682 | IQGAP2 | ENSG00000145703 | ENST00000502745 | -0.1024 | 565 | - | CHOL,KIRC,HNSC,KIRP,ESCA,LUSC,COAD | | ENSG00000280206 | ENST00000624682 | ACSL1 | ENSG00000151726 | ENST00000513317 | -0.1230 | 156 | - | CHOL,HNSC,KIRP,LUSC,LIHC | | ENSG00000280206 | ENST00000624682 | BTD | ENSG00000169814 | ENST00000449107 | -0.1884 | 815 | - | CHOL,ESCA,LUSC,COAD | | ENSG00000280206 | ENST00000624682 | BTD | ENSG00000169814 | ENST00000437172 | -0.1103 | 84 | - | CHOL,ESCA,LUSC,COAD | | ENSG00000280206 | ENST00000624682 | PC | ENSG00000173599 | ENST00000393955 | -0.1590 | 1 | - | GBM,CHOL,KIRC,KIRP,COAD | | ENSG00000280206 | ENST00000624682 | PC | ENSG00000173599 | ENST00000393958 | -0.1590 | 1 | - | GBM,CHOL,KIRC,KIRP,COAD | | ENSG00000280206 | ENST00000624682 | PC | ENSG00000173599 | ENST00000393960 | -0.1590 | 1 | - | GBM,CHOL,KIRC,KIRP,COAD | | ENSG00000280206 | ENST00000624682 | PC | ENSG00000173599 | ENST00000524491 | -0.2331 | 244 | - | GBM,CHOL,KIRC,KIRP,COAD | | ENSG00000280206 | ENST00000624682 | ABAT | ENSG00000183044 | ENST00000569156 | -0.1031 | 775 | - | CHOL,KIRC,KIRP,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | ABAT | ENSG00000183044 | ENST00000562115 | -0.1781 | 119 | - | CHOL,KIRC,KIRP,ESCA,LIHC | | ENSG00000280206 | ENST00000624682 | TMEM220 | ENSG00000187824 | ENST00000578345 | -0.2100 | 501 | - | CHOL,ESCA,LUSC,LUAD,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | TMEM220 | ENSG00000187824 | ENST00000581949 | -1.5767 | 501 | - | CHOL,ESCA,LUSC,LUAD,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | NKIRAS1 | ENSG00000197885 | ENST00000416026 | -0.7312 | 5 | - | GBM,CHOL | | ENSG00000280206 | ENST00000624682 | ETFRF1 | ENSG00000205707 | ENST00000554266 | -0.1091 | 565 | - | CHOL,LIHC | | ENSG00000280206 | ENST00000624682 | ARHGAP44 | ENSG00000006740 | ENST00000581437 | -0.2182 | 232 | - | LUSC,LUAD,COAD | | ENSG00000280206 | ENST00000624682 | NAAA | ENSG00000138744 | ENST00000513045 | -0.1269 | 395 | - | CHOL,HNSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | NAAA | ENSG00000138744 | ENST00000602782 | -0.1347 | 212 | - | CHOL,HNSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | MINDY1 | ENSG00000143409 | ENST00000493834 | -0.1089 | 303 | - | HNSC,ESCA,COAD | | ENSG00000280206 | ENST00000624682 | MINDY1 | ENSG00000143409 | ENST00000497067 | -0.1624 | 194 | - | HNSC,ESCA,COAD | | ENSG00000280206 | ENST00000624682 | CNNM2 | ENSG00000148842 | ENST00000369875 | -0.1141 | 318 | - | KIRC,KIRP,COAD | | ENSG00000280206 | ENST00000624682 | CITED2 | ENSG00000164442 | ENST00000618718 | -0.2098 | 212 | - | CHOL,HNSC,LUSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | LARS2 | ENSG00000011376 | ENST00000414984 | -0.2758 | 158 | - | CHOL,KIRC | | ENSG00000280206 | ENST00000624682 | MAPK9 | ENSG00000050748 | ENST00000347470 | -0.1072 | 706 | - | GBM | | ENSG00000280206 | ENST00000624682 | AP5M1 | ENSG00000053770 | ENST00000431972 | -1.5767 | 1 | - | KIRC | | ENSG00000280206 | ENST00000624682 | GALC | ENSG00000054983 | ENST00000393568 | -0.1408 | 543 | - | LUSC | | ENSG00000280206 | ENST00000624682 | GALC | ENSG00000054983 | ENST00000622264 | -1.2700 | 118 | - | LUSC | | ENSG00000280206 | ENST00000624682 | MGAT4A | ENSG00000071073 | ENST00000409391 | -0.3444 | 1 | - | HNSC,LUSC,COAD | | ENSG00000280206 | ENST00000624682 | WDR7 | ENSG00000091157 | ENST00000589935 | -0.1536 | 129 | - | GBM | | ENSG00000280206 | ENST00000624682 | NT5C2 | ENSG00000076685 | ENST00000552185 | -0.1006 | 30 | - | HNSC | | ENSG00000280206 | ENST00000624682 | NT5C2 | ENSG00000076685 | ENST00000470299 | -0.1840 | 331 | - | HNSC | | ENSG00000280206 | ENST00000624682 | MFAP3L | ENSG00000198948 | ENST00000506110 | -0.1527 | 355 | - | GBM,CHOL,KIRC,KIRP,LIHC | | ENSG00000280206 | ENST00000624682 | CERT1 | ENSG00000113163 | ENST00000261415 | -0.3381 | 459 | - | GBM,LUSC | | ENSG00000280206 | ENST00000624682 | ZNF106 | ENSG00000103994 | ENST00000569648 | -0.1049 | 156 | - | HNSC,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | CCSER2 | ENSG00000107771 | ENST00000359979 | -0.1002 | 372 | - | GBM | | ENSG00000280206 | ENST00000624682 | UHRF1BP1L | ENSG00000111647 | ENST00000548712 | -0.2112 | 1 | - | GBM,CHOL | | ENSG00000280206 | ENST00000624682 | KIAA1191 | ENSG00000122203 | ENST00000510164 | -0.1081 | 112 | - | KIRP,ESCA | | ENSG00000280206 | ENST00000624682 | KIAA1191 | ENSG00000122203 | ENST00000533553 | -0.1605 | 159 | - | KIRP,ESCA | | ENSG00000280206 | ENST00000624682 | BCL2L2 | ENSG00000129473 | ENST00000556599 | -0.2317 | 406 | - | GBM | | ENSG00000280206 | ENST00000624682 | IL6ST | ENSG00000134352 | ENST00000502326 | -0.6267 | 389 | - | CHOL,HNSC,ESCA,LUSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | TNS3 | ENSG00000136205 | ENST00000442536 | -0.2513 | 363 | - | KIRP,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | TNS3 | ENSG00000136205 | ENST00000458317 | -0.2448 | 373 | - | KIRP,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | TEX2 | ENSG00000136478 | ENST00000583501 | -0.1310 | 165 | - | GBM | | ENSG00000280206 | ENST00000624682 | TK2 | ENSG00000166548 | ENST00000525974 | -0.1091 | 155 | - | LUSC | | ENSG00000280206 | ENST00000624682 | TK2 | ENSG00000166548 | ENST00000417693 | -0.1098 | 1 | - | LUSC | | ENSG00000280206 | ENST00000624682 | TK2 | ENSG00000166548 | ENST00000563369 | -3.1800 | 96 | - | LUSC | | ENSG00000280206 | ENST00000624682 | TOM1L2 | ENSG00000175662 | ENST00000540946 | -0.1070 | 142 | - | GBM,KIRC,KIRP,ESCA,LUSC,LUAD,COAD | | ENSG00000280206 | ENST00000624682 | TOM1L2 | ENSG00000175662 | ENST00000542206 | -0.2455 | 169 | - | GBM,KIRC,KIRP,ESCA,LUSC,LUAD,COAD | | ENSG00000280206 | ENST00000624682 | SNX18 | ENSG00000178996 | ENST00000326277 | -0.1683 | 181 | - | CHOL,KIRP | | ENSG00000280206 | ENST00000624682 | CLMN | ENSG00000165959 | ENST00000556441 | -0.1779 | 360 | - | GBM,KIRC,KIRP,COAD | | ENSG00000280206 | ENST00000624682 | CLMN | ENSG00000165959 | ENST00000553733 | -0.1210 | 110 | - | GBM,KIRC,KIRP,COAD | | ENSG00000280206 | ENST00000624682 | TMPRSS2 | ENSG00000184012 | ENST00000458356 | -0.1121 | 163 | - | KIRC,HNSC,KIRP,LUSC,COAD,LIHC | | ENSG00000280206 | ENST00000624682 | CDKL5 | ENSG00000008086 | ENST00000379996 | -0.1546 | 170 | - | GBM,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | CDKL5 | ENSG00000008086 | ENST00000379989 | -0.1990 | 161 | - | GBM,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | EPS15 | ENSG00000085832 | ENST00000465467 | -0.1213 | 697 | - | GBM | | ENSG00000280206 | ENST00000624682 | CLIP4 | ENSG00000115295 | ENST00000401617 | -0.2146 | 667 | - | GBM | | ENSG00000280206 | ENST00000624682 | CLIP4 | ENSG00000115295 | ENST00000404424 | -0.5167 | 65 | - | GBM | | ENSG00000280206 | ENST00000624682 | REEP5 | ENSG00000129625 | ENST00000504247 | -0.2937 | 393 | - | GBM | | ENSG00000280206 | ENST00000624682 | BTBD9 | ENSG00000183826 | ENST00000419706 | -0.5292 | 179 | - | GBM,LUSC | | ENSG00000280206 | ENST00000624682 | CHP1 | ENSG00000187446 | ENST00000560397 | -0.6783 | 28 | - | GBM,CHOL,COAD | | ENSG00000280206 | ENST00000624682 | CDH1 | ENSG00000039068 | ENST00000422392 | -0.2817 | 173 | - | KIRC,KIRP | | ENSG00000280206 | ENST00000624682 | HACD3 | ENSG00000074696 | ENST00000565299 | -0.2485 | 131 | - | KIRC,KIRP | | ENSG00000280206 | ENST00000624682 | ILDR1 | ENSG00000145103 | ENST00000393631 | -1.0071 | 554 | - | KIRC | | ENSG00000280206 | ENST00000624682 | ILDR1 | ENSG00000145103 | ENST00000462014 | -0.4469 | 174 | - | KIRC | | ENSG00000280206 | ENST00000624682 | WDR72 | ENSG00000166415 | ENST00000559418 | -0.1426 | 1 | - | KIRC,KIRP,LIHC | | ENSG00000280206 | ENST00000624682 | PTPN4 | ENSG00000088179 | ENST00000431283 | -0.1521 | 317 | - | GBM,KIRC,KIRP | | ENSG00000280206 | ENST00000624682 | PTPN4 | ENSG00000088179 | ENST00000441089 | -0.5637 | 790 | - | GBM,KIRC,KIRP | | ENSG00000280206 | ENST00000624682 | ABLIM1 | ENSG00000099204 | ENST00000440467 | -0.1442 | 269 | - | GBM,KIRC,HNSC,ESCA | | ENSG00000280206 | ENST00000624682 | CPT1A | ENSG00000110090 | ENST00000539743 | -0.3172 | 97 | - | KIRP,COAD | | ENSG00000280206 | ENST00000624682 | RASSF8 | ENSG00000123094 | ENST00000539545 | -0.1394 | 58 | - | KIRC,KIRP,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | LMO7 | ENSG00000136153 | ENST00000526202 | -0.1235 | 1 | - | KIRC,HNSC,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | CRY2 | ENSG00000121671 | ENST00000616080 | -0.2292 | 335 | - | GBM,CHOL,HNSC,ESCA,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | GCC2 | ENSG00000135968 | ENST00000614209 | -0.6329 | 621 | - | GBM | | ENSG00000280206 | ENST00000624682 | APPL2 | ENSG00000136044 | ENST00000551662 | -0.1521 | 309 | - | COAD | | ENSG00000280206 | ENST00000624682 | SECISBP2L | ENSG00000138593 | ENST00000561428 | -0.1200 | 154 | - | GBM,HNSC,ESCA,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | SEC24B | ENSG00000138802 | ENST00000504968 | -0.2081 | 308 | - | CHOL,LIHC | | ENSG00000280206 | ENST00000624682 | SEC24B | ENSG00000138802 | ENST00000399100 | -0.2591 | 754 | - | CHOL,LIHC | | ENSG00000280206 | ENST00000624682 | FAM160B1 | ENSG00000151553 | ENST00000369246 | -0.8511 | 337 | - | GBM,CHOL | | ENSG00000280206 | ENST00000624682 | FCHO2 | ENSG00000157107 | ENST00000287761 | -0.1138 | 118 | - | HNSC,LUSC | | ENSG00000280206 | ENST00000624682 | ATP1A1 | ENSG00000163399 | ENST00000440951 | -0.1248 | 218 | - | KIRC,KIRP | | ENSG00000280206 | ENST00000624682 | DAG1 | ENSG00000173402 | ENST00000435508 | -0.3231 | 410 | - | KIRC | | ENSG00000280206 | ENST00000624682 | PCCA | ENSG00000175198 | ENST00000413170 | -0.1734 | 72 | - | CHOL,KIRC,HNSC,ESCA | | ENSG00000280206 | ENST00000624682 | ANKRD46 | ENSG00000186106 | ENST00000519316 | -0.3270 | 179 | - | GBM,CHOL,KIRP | | ENSG00000280206 | ENST00000624682 | LYRM7 | ENSG00000186687 | ENST00000507584 | -0.1466 | 320 | - | HNSC,KIRP | | ENSG00000280206 | ENST00000624682 | UTRN | ENSG00000152818 | ENST00000367526 | -0.5100 | 22 | - | KIRP,ESCA,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | PTPRM | ENSG00000173482 | ENST00000577468 | -0.1516 | 160 | - | GBM,ESCA,LUSC,LUAD | | ENSG00000280206 | ENST00000624682 | ACBD5 | ENSG00000107897 | ENST00000375888 | -0.1261 | 310 | - | CHOL | | ENSG00000280206 | ENST00000624682 | PLEKHB2 | ENSG00000115762 | ENST00000404460 | -0.1070 | 74 | - | GBM |
 lncRNA regulates mRNA by competing the miRNA binding site with mRNA. lncRNA regulates mRNA by competing the miRNA binding site with mRNA. |
| LncRNA Ensembl ID | lncRNA-miRNA-mRNA | Cancer Types | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PPIH | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPS27 | CHOL,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,WDR33 | CHOL,ACC,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NDE1 | CHOL,ACC,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,POLR2D | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CENPH | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DTYMK | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DPH7 | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,UCKL1 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ZNF48 | CHOL,MESO,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MUS81 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NRM | CHOL,ACC,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RXRB | CHOL,ACC,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PYCR2 | CHOL,UCS,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C1orf35 | CHOL,ACC,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CERS5 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DAXX | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CKS1B | CHOL,ACC,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ATAT1 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EFNA4 | CHOL,MESO,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FAM160B2 | CHOL,MESO,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SPOP | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RIT1 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,COIL | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM6 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DHX30 | CHOL,MESO,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,USP21 | CHOL,MESO,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TADA2A | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FUZ | CHOL,UCS,SARC,THYM,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PRR14 | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SF3B4 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C2orf68 | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM8A | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TONSL | CHOL,ACC,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NRF1 | CHOL,MESO,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DCAF15 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CCNF | CHOL,ACC,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBBP4 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PSMD4 | CHOL,ACC,MESO,UCS,THYM | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RING1 | CHOL,ACC,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ZNF7 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FANCE | CHOL,ACC,MESO,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,STX10 | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ILKAP | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SAMD1 | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NAA16 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FAM117B | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PRCC | CHOL,ACC,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ACYP1 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RCE1 | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TRAIP | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPS21 | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RSBN1 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM10 | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,JPT2 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KAT2A | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NCAPH2 | CHOL,ACC,MESO,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SNRPC | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LSM2 | CHOL,ACC,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DONSON | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,STK36 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SRSF3 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EHMT2 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SCMH1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ERCC3 | CHOL,ACC,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PHC1 | CHOL,MESO,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ISY1 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM4B | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,UBXN1 | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MYL6B | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EXOSC2 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LMNB2 | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RAD52 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LBR | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TOP1MT | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DDR1 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DNMT3A | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TRABD | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ASXL1 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C17orf75 | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CBX2 | CHOL,ACC,MESO,UCS,SARC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C2orf49 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SFI1 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,B3GALT6 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,H2AX | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DTL | CHOL,ACC,MESO,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TMEM201 | CHOL,ACC,MESO,UCS,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,POLR2H | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SALL2 | CHOL,MESO,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NAB1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM39 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TUBD1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PIDD1 | CHOL,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,GNL1 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MBD1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CBFB | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TNRC18 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,BRAT1 | CHOL,MESO,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FAM111B | CHOL,ACC,MESO,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SLC29A2 | CHOL,MESO,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KDM1A | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ING5 | CHOL,ACC,MESO,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SAP30BP | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CEP131 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CLK2 | CHOL,ACC,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C1orf159 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SSRP1 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CCDC77 | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RAD9A | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MCM7 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PRPF38A | CHOL,ACC,MESO,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LUC7L2 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,POLE | CHOL,ACC,MESO,UCS,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KIFC1 | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SMPD4 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,HDAC1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KHDC4 | CHOL,ACC,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PHF10 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ANP32A | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CHD1L | CHOL,ACC,MESO,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MRPL53 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CDKN2C | ACC,MESO,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DHX34 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NSMCE2 | CHOL,ACC,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TIA1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SLC41A3 | CHOL,MESO,UCS,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TLE2 | CHOL,MESO,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NOC2L | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DPF2 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PTMA | CHOL,ACC,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ARFIP2 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NOP58 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,GABBR1 | CHOL,MESO,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,STMN1 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PNKP | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DDX11 | CHOL,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,BANF1 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SBK1 | CHOL,MESO,SARC,THYM,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TSEN54 | CHOL,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TGIF2 | CHOL,ACC,MESO,UCS,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FLVCR1 | CHOL,MESO,UCS,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ATXN7L2 | CHOL,ACC,UCS,THYM,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TFPT | CHOL,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SIVA1 | ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CRTC2 | CHOL,MESO,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CDC25C | CHOL,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ASF1B | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CDCA5 | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SMG5 | CHOL,ACC,UCS,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TMEM44 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,E2F1 | CHOL,MESO,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SF3A3 | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NVL | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PPP1R26 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPL37A | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,AKAP8 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SMARCC1 | CHOL,ACC,MESO,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBMX | CHOL,ACC,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CACNB3 | CHOL,ACC,MESO,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RFC2 | CHOL,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,H3-3B | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,IQCC | CHOL,UCS,SARC,THYM,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DGUOK | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PMS1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EZH2 | CHOL,ACC,MESO,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FAM136A | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KIF22 | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,UTP6 | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FBXO46 | CHOL,ACC,MESO,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ATG4B | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CELF1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MSL2 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPS27A | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPS10 | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM5 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CCDC57 | CHOL,UCS,SARC,THYM,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SNRPA | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ARL16 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TCF19 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CXXC1 | CHOL,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CHAF1B | CHOL,ACC,MESO,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,COPS7B | CHOL,ACC,MESO,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPL36A | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,INTS11 | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CHTF18 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,HROB | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CPSF7 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SUMO2 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,HNRNPUL2 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PTK7 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MRPL11 | CHOL,ACC,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MRE11 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EPN2 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TROAP | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PHIP | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,USF1 | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C14orf93 | CHOL,ACC,MESO,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SYMPK | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KMT5C | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RMI2 | CHOL,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CLASP1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CHTOP | CHOL,ACC,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SPC25 | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM14 | CHOL,ACC,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FAF1 | CHOL,ACC,MESO,UCS,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,AP4M1 | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CLASRP | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,HINFP | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CDC37 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,UBTF | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MKS1 | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PCIF1 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NASP | CHOL,ACC,MESO,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PURB | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C11orf80 | CHOL,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ABI2 | CHOL,ACC,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FNBP4 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ANP32B | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TFAP4 | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SRSF1 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,DGCR8 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,BCL2L12 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,POGZ | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,UBXN11 | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,BRD8 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,IRF3 | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,THOC2 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CHEK1 | CHOL,ACC,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FANCG | CHOL,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPS11 | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TEDC2 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPL35A | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,BUD13 | CHOL,ACC,MESO,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM22 | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SSBP4 | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ATAD3B | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PRR3 | CHOL,ACC,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NUP85 | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,IQCB1 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KDM5B | CHOL,MESO,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CDCA7L | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MUTYH | CHOL,ACC,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LYSMD1 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,HMGB2 | CHOL,ACC,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ING4 | CHOL,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PRKRA | CHOL,ACC,UCS,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RCCD1 | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MCM2 | CHOL,ACC,MESO,UCS,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FEN1 | CHOL,MESO,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CENPJ | CHOL,ACC,MESO,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RGL3 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,AGO2 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LIG1 | CHOL,MESO,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NT5C | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,C11orf49 | CHOL,ACC,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SRRT | CHOL,ACC,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PMF1 | CHOL,ACC,UCS,THYM,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,YY1AP1 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TMEM216 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SF3A2 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CASC3 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ZMYM3 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SPINDOC | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TTC13 | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,H3-3A | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LRRC14 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RSL24D1 | CHOL,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TAF1D | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CDC45 | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MTA1 | CHOL,ACC,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SCNM1 | CHOL,ACC,MESO,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,PGAP2 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,LSM7 | CHOL,ACC,MESO,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,BRD4 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TMEM39B | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EIF3F | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ANP32E | CHOL,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NFRKB | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RFC4 | CHOL,ACC,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RBM33 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RRP8 | CHOL,ACC,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NCK2 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,ILF3 | CHOL,ACC,MESO,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,TYMS | CHOL,ACC,MESO,SARC,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,MCM5 | CHOL,ACC,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EIF3H | CHOL,ACC,SARC,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CCAR2 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,NELFE | CHOL,ACC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,HDGFL2 | CHOL,MESO,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SZT2 | CHOL,ACC,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CCDC14 | CHOL,UCS,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPS29 | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,POLD1 | CHOL,ACC,MESO,UCS,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,GTF2H4 | CHOL,ACC,UCS,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,EDC3 | CHOL,ACC,MESO,SARC,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,KIF2C | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,YJEFN3 | CHOL,ACC,MESO,SARC,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,FBXL12 | CHOL,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,RPS8 | CHOL,ACC,UCS,THYM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,SKA2 | CHOL,ACC,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-22,CCHCR1 | CHOL,ACC,MESO,UCS,SARC,THYM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PRKRIP1 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,C1orf159 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CBFB | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,LSM5 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,RFC2 | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MIS18A | HNSC,CHOL,LGG,GBM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,SP1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NVL | CHOL,LGG,GBM,UCS,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,JPT2 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,FAAP100 | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PHF12 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,SIRT7 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,AP5Z1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CDCA7L | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,BICRA | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,TRA2A | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,SAP30BP | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,TARBP2 | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,RALY | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PGLS | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MGME1 | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,GTF3C5 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,C1orf35 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,C1QTNF6 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,EMC8 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,UBALD2 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NKIRAS2 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,ACTL6A | HNSC,CHOL,LGG,GBM,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,RNPEPL1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,POFUT2 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NELFA | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PPIE | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CBX3 | HNSC,CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MOGS | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CCDC130 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NRM | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,WDR90 | CHOL,LGG,GBM,UCS,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MITD1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MEX3D | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,METTL17 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,KCTD11 | CHOL,LGG,GBM,UCS,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CXorf38 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CDK2AP2 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,GIGYF1 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,UBE2Z | HNSC,CHOL,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CYREN | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PDAP1 | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PLOD3 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,POLD1 | HNSC,CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,RHNO1 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,XRCC1 | HNSC,CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,C14orf93 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MKS1 | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,DTYMK | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NASP | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MUTYH | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NDE1 | HNSC,CHOL,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PTBP1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,AUP1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NUB1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,TRIM56 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PFN1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,FBRS | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,SLC12A9 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,TPD52L2 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,KRT10 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,POMGNT1 | CHOL,LGG,GBM,UCS,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,IST1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PELP1 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,DTX2 | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PCIF1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,LEMD2 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CLK2 | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,SNRPA | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,KMT5C | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,IRF3 | CHOL,LGG,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CEP95 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,P3H1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,C1orf131 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,SOX9 | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PSMC3IP | HNSC,CHOL,LGG,GBM,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,IFT52 | HNSC,CHOL,LGG,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,NOL4L | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MFSD10 | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,BAX | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,EFNA1 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CDK2 | HNSC,CHOL,LGG,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,PPIH | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,MCM3 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,DHX34 | CHOL,LGG,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,POP5 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,ATRAID | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,ARL6IP6 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,RBM39 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,KXD1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,OFD1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,GINS2 | HNSC,CHOL,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,DDR1 | CHOL,LGG,GBM,UCS,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,STX10 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,ZNF3 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,EIF2B1 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CARD19 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,CLN3 | CHOL,LGG,GBM,UCS,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,DNAAF5 | CHOL,LGG,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-139,TRAF2 | CHOL,GBM,UCS,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,CNOT11 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,PRKRA | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,EHBP1L1 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,PML | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,SEMA4C | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,YEATS2 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,NMI | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,TRAF4 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,VMP1 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,ADGRE5 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,CASP8 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,ARPC1B | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,PIAS3 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,ZYX | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,SLC39A1 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,NDE1 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,AGTRAP | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,JPT1 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,LYPLA2 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,RNF149 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,ANXA2 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,FAM136A | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,PPM1G | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,SFR1 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,ARPC2 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-1468,ITGB5 | KICH,CHOL,ACC,PAAD,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,CHEK1 | KICH,ACC,GBM,MESO,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,MRGBP | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,RBM28 | KICH,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,PLOD3 | KICH,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,CHTF18 | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,AGPS | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,BMP1 | KICH,ACC,GBM,MESO,KIRP | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,MCM7 | KICH,ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,HMBS | KICH,ACC,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,AURKA | KICH,ACC,GBM,MESO,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,PIDD1 | KICH,ACC,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,POLD1 | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,RALA | KICH,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,SAAL1 | KICH,ACC,GBM,MESO,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,LIG1 | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,KNOP1 | KICH,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,CHAF1A | KICH,ACC,GBM,MESO,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,SMPD4 | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,COPS7B | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,POGLUT2 | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,DTYMK | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,MYO19 | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,GINS2 | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,NCAPD2 | KICH,ACC,GBM,MESO,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,MCM3 | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,NRM | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,KMT5C | KICH,ACC,GBM,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,NASP | ACC,GBM,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,DONSON | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,FEN1 | KICH,ACC,GBM,MESO,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,CCDC88A | KICH,ACC,GBM,MESO,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,RFC2 | KICH,ACC,MESO,KIRP,LIHC | | ENSG00000280206 | CTB-193M12.5,hsa-mir-29c,TARBP2 | ACC,GBM,MESO,KIRP,LIHC |
 ORFfinder result for the gencode.v22.lncRNA.transcript.fa. ORFfinder result for the gencode.v22.lncRNA.transcript.fa. |
| lncRNA Ensembl ID | lncRNA ENST ID | length(AA) | start at transcript | end at transcript | | ENSG00000280206.1 | ENST00000624682.1 | 96 | 213 | 500 |
|

 Gene summary
Gene summary AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure.
AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure.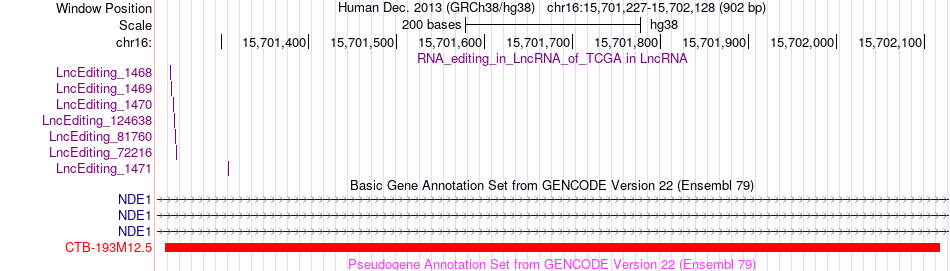
 Differentially expressed gene analysis between cancer and normal samples.
Differentially expressed gene analysis between cancer and normal samples. Correlation between lncRNAs and cancer stages.
Correlation between lncRNAs and cancer stages.
 lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5).
lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5). lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5).
lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5). lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5).
lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5).







