 lncRNA targets the enhancer region and positively regulates the gene expression. lncRNA targets the enhancer region and positively regulates the gene expression. |
| Only predicted by TDF | | ZNF30,CEBPZ,PUM1,XRN1,KLF9,ILF3,NOL8,PBX2,RBM39,ZBTB4,KLF7,CIRBP,LCORL,JAZF1,MZF1,MYSM1,NRF1,CSDE1,RBM23,AKAP8,ZNF2,ARID2,ZXDC,SUGP2,RARB,DMTF1,VEZF1,RBM27,ZNF70,BAZ2A,ZNF77,ZNF24,RORA,ZNF74,TBP,GABPA,NFKB1,IKZF5,ZBED4,MEF2C,DDX55,KMT2B,TET3,ZBTB5,ZFP14,ZXDA,RNPC3,ZFP69,SAFB2,NOP9,THOC1,ZNF25,FOXO1,LARP7,PNPT1,CNOT4,SON,CHTOP,CWC22,DDX59,ZZZ3,ZBED5,SCAF4,CENPT,TTC14,ZBED3,RBM4,SMAD9,CREB1,MEX3C,RC3H1,ZNF44,ZNF91,ZFP91,RBMX,AGO2,RBM4B,U2AF2,CSTF3,RBMS3,TIGD6,RBM26,ZFP1,TCFL5,ZIK1,RBM14,PURB,GZF1,ADNP,NCBP1,MYEF2,PPIL4,TERF2,MBD1,IKZF4,PUS7L,ASH1L,CLOCK,CASC3,FUBP1,RBM15,ARNT2,ZFP30,SP4,RBM43,CPSF7,KLF11,PRDM4,TCF20,LIN54,AGFG1,UBR5,THAP9,THOC2,TSN,ZBTB6,CXXC1,ZNF34,ZNF71,THAP6,DDX17,E2F5,ZBTB2,SRRM2,RFX7,E2F6,FOXN3,AQR,ZNF22,ZHX3,FMR1,MECP2,LCOR,FOXJ3,SRSF9,KLF3,PURA,ZNF41,KLF15,ZC3H8,TTF1,RBM41,SART3,THAP1,NOL9,DZIP3,SRBD1,ILF2,ZFP62,ZNF93,PAIP1,ZNF28,ZNF45,CELF1,SCMH1,PSPC1,KRR1,TMF1,ELK4,TIGD1,RBM45,NFXL1,PRDM2,DDX51,MBD4,RBM18,UNC50,NFIB,SRSF2,MBD6,DDX31,SALL2,MYNN,TFCP2,ZMAT1,ZNF83,QKI,ZFR,NSUN6,ZNF3,NXF1,ZFX,ZNF8,NR2C2,EIF2D,SYNJ1,GPBP1,C1D,ARNT,RBM6,HMGN3,ZNF14,G3BP2,RBM48,LSM11,DZIP1,PUM2,XPO1,RBM5,ELF2,SMAD4,RBMS1,NFYB,ZNF43,ZFP28,MTF1,SP3,ZNF17,SRCAP,DOT1L,ZNF84,DDX42,BAZ2B,RXRB,TRA2A,RBM8A,KMT2A,ZEB2,ZNF69,SRSF7,HINFP,XPA,AKAP1,NR2C1,MAF,TFDP2,ZFP3,TEP1,SFPQ,TET2,RBM33,ATF2,ZNF10 | | Exist in public source | | NA |
 lncRNA targets the promoter region and positively regulates the gene expression. lncRNA targets the promoter region and positively regulates the gene expression. |
 lncRNA targets the promoter region and negatively regulates the gene expression. lncRNA targets the promoter region and negatively regulates the gene expression. |
 lncRNA targets the 3'UTR region and negatively regulates the mRNA. lncRNA targets the 3'UTR region and negatively regulates the mRNA. |
| Only predicted by lncTar | | TEAD4 | | Exist in public source | | NA |
 lncRNA targets the skipped exon region. lncRNA targets the skipped exon region. | | -lncRNA and exonskipping events are positively correlated. |
| LncRNA Ensembl ID | LncRNA ENST ID | Exon ID | Skipped Exon | dG | ndG | Gene name with skipped exon | TransID with skipped exon | ORF mutation | | ENSG00000263753 | ENST00000577439 | exon_skip_460899 | chr6:85538837-85538864 | -5.77 | -0.3037 | SNX14 | ENST00000314673 | In-frame | | ENSG00000263753 | ENST00000577439 | exon_skip_150854 | chr17:32366664-32366757 | -11.00 | -0.2973 | ZNF207 | ENST00000321233 | In-frame |
| -lncRNA and exonskipping events are negatively correlated. |
| LncRNA Ensembl ID | LncRNA ENST ID | Exon ID | Skipped Exon | dG | ndG | Gene name with skipped exon | TransID with skipped exon | LOF | | ENSG00000263753 | ENST00000577439 | exon_skip_139422 | chr16:89114338-89114487 | -19.85 | -0.1378 | ACSF3 | ENST00000317447,ENST00000406948 | Frame-shift | | ENSG00000263753 | ENST00000577439 | exon_skip_361485 | chr21:29008239-29008307 | -10.96 | -0.1858 | RWDD2B | ENST00000493196 | Frame-shift | | ENSG00000263753 | ENST00000577439 | exon_skip_382458 | chr3:38131595-38131638 | -9.84 | -0.2659 | ACAA1 | ENST00000333167 | Frame-shift |
 lncRNA targets by miRNA. lncRNA targets by miRNA. |
 |
| LncRNA Ensembl ID | miRNA ID | LncRNA ENST ID | Binding site in lncRNA | Score | Energy | Align Len | Public source | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5238308-5238428 | 240.00 | -99.59 | 118 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5239325-5239432 | 217.00 | -86.69 | 110 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5238618-5238742 | 209.00 | -92.26 | 122 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5237858-5237982 | 197.00 | -100.44 | 121 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5238901-5239005 | 197.00 | -76.52 | 111 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5238142-5238256 | 194.00 | -104.27 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5238461-5238573 | 194.00 | -118.20 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5238051-5238152 | 179.00 | -112.04 | 107 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5239545-5239663 | 170.00 | -99.41 | 119 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5239069-5239200 | 168.00 | -91.27 | 125 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5238778-5238901 | 167.00 | -99.22 | 116 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5239685-5239794 | 154.00 | -86.94 | 107 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5239203-5239314 | 150.00 | -85.48 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000581471 | chr18:5239454-5239549 | 144.00 | -77.90 | 101 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5238308-5238428 | 240.00 | -99.59 | 118 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5239309-5239425 | 222.00 | -82.80 | 114 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5238618-5238742 | 209.00 | -92.26 | 122 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5238954-5239066 | 208.00 | -91.51 | 115 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5239452-5239574 | 202.00 | -86.02 | 110 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5239739-5239846 | 199.00 | -99.43 | 110 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5238142-5238256 | 194.00 | -104.27 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5238461-5238573 | 194.00 | -118.20 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5240591-5240716 | 194.00 | -93.03 | 123 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5240053-5240164 | 191.00 | -79.04 | 110 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5239109-5239229 | 186.00 | -82.50 | 101 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5239949-5240056 | 186.00 | -102.05 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5240869-5240974 | 186.00 | -96.13 | 111 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5240383-5240493 | 184.00 | -80.75 | 95 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5240708-5240828 | 179.00 | -97.45 | 120 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5240209-5240319 | 178.00 | -87.29 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5239581-5239695 | 169.00 | -88.28 | 113 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5238778-5238901 | 167.00 | -99.22 | 116 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582363 | chr18:5238085-5238152 | 141.00 | -87.63 | 51 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5238308-5238428 | 240.00 | -99.59 | 118 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5241724-5241830 | 229.00 | -103.96 | 114 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5239824-5239936 | 214.00 | -88.26 | 106 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5238618-5238742 | 209.00 | -92.26 | 122 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5240493-5240623 | 208.00 | -93.31 | 120 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5240782-5240895 | 208.00 | -94.30 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5241916-5242017 | 205.00 | -88.45 | 105 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5241607-5241726 | 198.00 | -96.39 | 115 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5238901-5239005 | 197.00 | -76.52 | 111 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5238142-5238256 | 194.00 | -104.27 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5238461-5238573 | 194.00 | -118.20 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5239079-5239204 | 194.00 | -93.03 | 123 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5241003-5241120 | 191.00 | -94.15 | 117 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5239357-5239462 | 186.00 | -96.13 | 111 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5241322-5241446 | 183.00 | -93.06 | 123 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5240295-5240409 | 182.00 | -100.28 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5241200-5241314 | 180.00 | -81.97 | 109 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5239196-5239316 | 179.00 | -97.45 | 120 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5240160-5240259 | 179.00 | -87.14 | 109 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5239537-5239650 | 176.00 | -83.85 | 114 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5240670-5240774 | 174.00 | -86.59 | 106 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5240005-5240109 | 172.00 | -97.78 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5238778-5238901 | 167.00 | -99.22 | 116 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5240892-5241011 | 158.00 | -88.03 | 116 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5239734-5239842 | 156.00 | -80.62 | 98 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5239640-5239740 | 155.00 | -75.31 | 104 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5238101-5238152 | 141.00 | -77.90 | 51 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000582008 | chr18:5241488-5241597 | 140.00 | -103.25 | 106 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580082 | chr18:5238308-5238428 | 240.00 | -99.59 | 118 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580082 | chr18:5238618-5238742 | 209.00 | -92.26 | 122 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580082 | chr18:5238142-5238256 | 194.00 | -104.27 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580082 | chr18:5238461-5238573 | 194.00 | -118.20 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580082 | chr18:5238849-5238957 | 185.00 | -94.79 | 105 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580684 | chr18:5238618-5238742 | 209.00 | -92.26 | 122 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580684 | chr18:5238901-5239005 | 197.00 | -76.52 | 111 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580684 | chr18:5239137-5239236 | 186.00 | -92.15 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580684 | chr18:5239434-5239539 | 186.00 | -96.13 | 111 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580684 | chr18:5239273-5239393 | 179.00 | -97.45 | 120 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580684 | chr18:5239019-5239119 | 175.00 | -85.57 | 105 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000580684 | chr18:5238778-5238901 | 167.00 | -99.22 | 116 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5244007-5244113 | 229.00 | -103.96 | 114 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5242107-5242219 | 214.00 | -88.26 | 106 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5242776-5242906 | 208.00 | -93.31 | 120 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5243065-5243178 | 208.00 | -94.30 | 112 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5244199-5244300 | 205.00 | -88.45 | 105 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5241215-5241325 | 198.00 | -78.51 | 107 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5243890-5244009 | 198.00 | -96.39 | 115 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5243286-5243403 | 191.00 | -94.15 | 117 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5241086-5241193 | 186.00 | -102.05 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5241343-5241442 | 186.00 | -92.15 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5241640-5241745 | 186.00 | -96.13 | 111 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5243605-5243729 | 183.00 | -93.06 | 123 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5242578-5242692 | 182.00 | -100.28 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5243483-5243597 | 180.00 | -81.97 | 109 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5241479-5241599 | 179.00 | -97.45 | 120 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5242443-5242542 | 179.00 | -87.14 | 109 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5241820-5241933 | 176.00 | -83.85 | 114 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5242953-5243057 | 174.00 | -86.59 | 106 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5242288-5242392 | 172.00 | -97.78 | 108 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5243175-5243294 | 158.00 | -88.03 | 116 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5242017-5242125 | 156.00 | -80.62 | 98 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5241923-5242023 | 155.00 | -75.31 | 104 | NA | | ENSG00000263753 | hsa-mir-34a | ENST00000577439 | chr18:5243771-5243880 | 140.00 | -103.25 | 106 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5238386-5238481 | 200.00 | -86.35 | 95 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5239422-5239509 | 188.00 | -69.91 | 90 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5238111-5238194 | 179.00 | -83.47 | 61 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5238234-5238315 | 168.00 | -87.49 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5237876-5237956 | 166.00 | -67.71 | 70 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5238619-5238706 | 164.00 | -69.99 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5238495-5238581 | 163.00 | -94.51 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5239087-5239166 | 161.00 | -67.89 | 72 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5239257-5239334 | 155.00 | -62.84 | 75 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5238787-5238876 | 154.00 | -69.57 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000581471 | chr18:5238053-5238134 | 153.00 | -95.19 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5238386-5238481 | 200.00 | -86.35 | 95 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239780-5239858 | 182.00 | -74.75 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5238111-5238194 | 179.00 | -83.47 | 61 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239283-5239375 | 179.00 | -65.56 | 81 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5240341-5240429 | 177.00 | -67.17 | 89 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5240155-5240245 | 175.00 | -73.92 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5238234-5238315 | 168.00 | -87.49 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5240676-5240755 | 166.00 | -65.10 | 70 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5238619-5238706 | 164.00 | -69.99 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5240836-5240922 | 164.00 | -73.57 | 79 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5238495-5238581 | 163.00 | -94.51 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239443-5239530 | 163.00 | -69.97 | 87 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5240527-5240613 | 163.00 | -57.61 | 81 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239931-5240011 | 161.00 | -79.70 | 68 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239156-5239242 | 160.00 | -65.45 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239618-5239700 | 160.00 | -71.87 | 85 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239083-5239164 | 158.00 | -74.02 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5238787-5238876 | 154.00 | -69.57 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5238979-5239067 | 154.00 | -80.84 | 81 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5240939-5241019 | 152.00 | -76.73 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582363 | chr18:5239861-5239951 | 151.00 | -76.11 | 89 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5238386-5238481 | 200.00 | -86.35 | 95 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5241603-5241680 | 198.00 | -66.67 | 75 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5240327-5240413 | 187.00 | -77.81 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5241694-5241783 | 181.00 | -71.69 | 86 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5238111-5238194 | 179.00 | -83.47 | 61 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5241021-5241113 | 178.00 | -70.12 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5239013-5239101 | 169.00 | -77.29 | 90 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5238234-5238315 | 168.00 | -87.49 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5239164-5239243 | 166.00 | -65.10 | 70 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5239794-5239871 | 166.00 | -62.87 | 81 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5240582-5240661 | 166.00 | -66.59 | 73 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5241327-5241420 | 166.00 | -73.44 | 87 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5238619-5238706 | 164.00 | -69.99 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5239324-5239410 | 164.00 | -73.57 | 79 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5238495-5238581 | 163.00 | -94.51 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5240692-5240773 | 157.00 | -72.89 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5241781-5241876 | 157.00 | -75.45 | 89 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5238787-5238876 | 154.00 | -69.57 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5239427-5239508 | 152.00 | -76.96 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5239687-5239776 | 149.00 | -66.90 | 90 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5240020-5240091 | 149.00 | -70.03 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5240168-5240265 | 148.00 | -69.93 | 95 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5240867-5240949 | 147.00 | -78.99 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5241878-5241963 | 144.00 | -72.23 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5241092-5241178 | 143.00 | -67.31 | 65 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000582008 | chr18:5239875-5239966 | 140.00 | -77.22 | 90 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580082 | chr18:5238386-5238481 | 200.00 | -86.35 | 95 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580082 | chr18:5238831-5238913 | 183.00 | -70.21 | 79 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580082 | chr18:5238129-5238194 | 179.00 | -69.00 | 61 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580082 | chr18:5238234-5238315 | 168.00 | -87.49 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580082 | chr18:5238619-5238706 | 164.00 | -69.99 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580082 | chr18:5238495-5238581 | 163.00 | -94.51 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580684 | chr18:5239139-5239228 | 178.00 | -71.54 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580684 | chr18:5239013-5239101 | 169.00 | -77.29 | 90 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580684 | chr18:5239241-5239320 | 166.00 | -65.10 | 70 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580684 | chr18:5238619-5238706 | 164.00 | -69.99 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580684 | chr18:5239401-5239487 | 164.00 | -73.57 | 79 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580684 | chr18:5238551-5238619 | 154.00 | -82.64 | 72 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000580684 | chr18:5238787-5238876 | 154.00 | -69.57 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5243886-5243963 | 198.00 | -66.67 | 75 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5242610-5242696 | 187.00 | -77.81 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5241220-5241307 | 185.00 | -61.42 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5243977-5244066 | 181.00 | -71.69 | 86 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5241345-5241434 | 178.00 | -71.54 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5243304-5243396 | 178.00 | -70.12 | 84 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5241447-5241526 | 166.00 | -65.10 | 70 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5242077-5242154 | 166.00 | -62.87 | 81 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5242865-5242944 | 166.00 | -66.59 | 73 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5243610-5243703 | 166.00 | -73.44 | 87 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5241607-5241693 | 164.00 | -73.57 | 79 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5241068-5241148 | 161.00 | -79.70 | 68 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5242975-5243056 | 157.00 | -72.89 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5244064-5244159 | 157.00 | -75.45 | 89 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5241710-5241791 | 152.00 | -76.96 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5240998-5241088 | 151.00 | -76.11 | 89 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5241970-5242059 | 149.00 | -66.90 | 90 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5242303-5242374 | 149.00 | -70.03 | 83 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5242451-5242548 | 148.00 | -69.93 | 95 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5243150-5243232 | 147.00 | -78.99 | 82 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5244161-5244246 | 144.00 | -72.23 | 78 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5243375-5243461 | 143.00 | -67.31 | 65 | NA | | ENSG00000263753 | hsa-mir-22 | ENST00000577439 | chr18:5242158-5242249 | 140.00 | -77.22 | 90 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5238850-5238962 | 201.00 | -82.14 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5238388-5238516 | 198.00 | -109.67 | 128 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5239607-5239726 | 195.00 | -79.52 | 107 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5238957-5239078 | 190.00 | -86.39 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5239168-5239277 | 189.00 | -77.68 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5239396-5239499 | 179.00 | -80.55 | 111 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5238177-5238287 | 176.00 | -104.73 | 99 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5239500-5239605 | 173.00 | -84.93 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5238521-5238641 | 170.00 | -109.57 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5238680-5238795 | 168.00 | -91.64 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5238044-5238161 | 163.00 | -107.32 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5239740-5239852 | 163.00 | -85.21 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5237928-5238054 | 161.00 | -112.00 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000581471 | chr18:5237827-5237912 | 145.00 | -76.11 | 91 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5240886-5241000 | 220.00 | -100.58 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5239071-5239192 | 209.00 | -87.91 | 119 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5240281-5240401 | 205.00 | -76.87 | 120 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5238850-5238962 | 201.00 | -82.14 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5238388-5238516 | 198.00 | -109.67 | 128 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5240165-5240283 | 197.00 | -82.93 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5240558-5240670 | 195.00 | -78.47 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5239582-5239690 | 194.00 | -75.34 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5239391-5239500 | 187.00 | -76.70 | 111 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5238957-5239080 | 185.00 | -94.57 | 119 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5239849-5239965 | 184.00 | -89.44 | 114 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5239218-5239356 | 179.00 | -98.75 | 136 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5238177-5238287 | 176.00 | -104.73 | 99 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5238521-5238641 | 170.00 | -109.57 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5238680-5238795 | 168.00 | -91.64 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5239751-5239850 | 168.00 | -86.81 | 111 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5240725-5240838 | 167.00 | -94.01 | 112 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5239968-5240087 | 160.00 | -102.06 | 99 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5240450-5240569 | 159.00 | -68.87 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582363 | chr18:5238085-5238182 | 149.00 | -93.22 | 95 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5239374-5239488 | 220.00 | -100.58 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5241656-5241773 | 206.00 | -89.28 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5238850-5238962 | 201.00 | -82.14 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5239045-5239158 | 200.00 | -75.28 | 113 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5239902-5240011 | 200.00 | -78.57 | 103 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5241008-5241127 | 200.00 | -89.52 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5238388-5238516 | 198.00 | -109.67 | 128 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5240011-5240114 | 197.00 | -86.82 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5241922-5242021 | 195.00 | -82.23 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5239740-5239851 | 188.00 | -79.23 | 114 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5240279-5240394 | 181.00 | -94.13 | 113 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5240801-5240907 | 179.00 | -86.47 | 108 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5238177-5238287 | 176.00 | -104.73 | 99 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5241485-5241605 | 175.00 | -96.99 | 109 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5241809-5241915 | 175.00 | -74.70 | 101 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5238521-5238641 | 170.00 | -109.57 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5240666-5240783 | 170.00 | -83.61 | 119 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5241147-5241265 | 170.00 | -87.36 | 117 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5241334-5241434 | 169.00 | -76.90 | 107 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5238680-5238795 | 168.00 | -91.64 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5239213-5239326 | 167.00 | -94.01 | 112 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5239598-5239704 | 166.00 | -68.14 | 108 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5240451-5240555 | 166.00 | -67.87 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5240178-5240282 | 151.00 | -78.07 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000582008 | chr18:5238101-5238161 | 141.00 | -74.91 | 59 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580082 | chr18:5238388-5238516 | 198.00 | -109.67 | 128 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580082 | chr18:5238767-5238873 | 195.00 | -92.68 | 107 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580082 | chr18:5238177-5238287 | 176.00 | -104.73 | 99 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580082 | chr18:5238521-5238641 | 170.00 | -109.57 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580082 | chr18:5238676-5238779 | 153.00 | -79.51 | 108 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580684 | chr18:5238850-5238962 | 201.00 | -82.14 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580684 | chr18:5239045-5239158 | 200.00 | -75.28 | 113 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580684 | chr18:5239233-5239346 | 172.00 | -82.45 | 109 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580684 | chr18:5239451-5239559 | 172.00 | -94.82 | 112 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580684 | chr18:5238680-5238795 | 168.00 | -91.64 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580684 | chr18:5238551-5238659 | 156.00 | -100.27 | 107 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000580684 | chr18:5239350-5239452 | 155.00 | -86.64 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5241657-5241771 | 220.00 | -100.58 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5243939-5244056 | 206.00 | -89.28 | 116 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5242185-5242294 | 200.00 | -78.57 | 103 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5243291-5243410 | 200.00 | -89.52 | 115 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5242294-5242397 | 197.00 | -86.82 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5241252-5241364 | 195.00 | -78.47 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5244205-5244304 | 195.00 | -82.23 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5242023-5242134 | 188.00 | -79.23 | 114 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5240991-5241102 | 184.00 | -88.06 | 114 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5242562-5242677 | 181.00 | -94.13 | 113 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5243084-5243190 | 179.00 | -86.47 | 108 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5243768-5243888 | 175.00 | -96.99 | 109 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5244092-5244198 | 175.00 | -74.70 | 101 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5241439-5241552 | 172.00 | -82.45 | 109 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5242949-5243066 | 170.00 | -83.61 | 119 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5243430-5243548 | 170.00 | -87.36 | 117 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5243617-5243717 | 169.00 | -76.90 | 107 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5241881-5241987 | 166.00 | -68.14 | 108 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5242734-5242838 | 166.00 | -67.87 | 110 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5241105-5241224 | 160.00 | -102.06 | 99 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5241556-5241658 | 155.00 | -86.64 | 106 | NA | | ENSG00000263753 | hsa-mir-221 | ENST00000577439 | chr18:5242461-5242565 | 151.00 | -78.07 | 106 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5238599-5238708 | 218.00 | -93.01 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5237973-5238081 | 210.00 | -122.24 | 104 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5238155-5238263 | 205.00 | -103.78 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5238402-5238510 | 204.00 | -108.20 | 107 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5238985-5239105 | 198.00 | -96.42 | 118 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5238728-5238837 | 189.00 | -94.34 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5239751-5239858 | 184.00 | -94.65 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5239144-5239268 | 179.00 | -92.42 | 122 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5239393-5239508 | 175.00 | -94.20 | 116 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5239535-5239646 | 164.00 | -101.20 | 110 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5238847-5238950 | 163.00 | -92.08 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5237867-5237980 | 156.00 | -97.68 | 107 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000581471 | chr18:5238299-5238414 | 156.00 | -94.28 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5238599-5238708 | 218.00 | -93.01 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5240892-5240992 | 217.00 | -100.28 | 96 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5238155-5238263 | 205.00 | -103.78 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5238402-5238510 | 204.00 | -108.20 | 107 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5240668-5240771 | 201.00 | -86.24 | 110 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5240389-5240491 | 197.00 | -82.54 | 109 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5238963-5239072 | 195.00 | -96.38 | 105 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5238728-5238837 | 189.00 | -94.34 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5239972-5240084 | 188.00 | -104.54 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5239675-5239795 | 186.00 | -97.29 | 115 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5239503-5239612 | 185.00 | -86.80 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5240148-5240270 | 184.00 | -95.69 | 122 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5239117-5239235 | 183.00 | -91.24 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5239839-5239961 | 181.00 | -99.97 | 114 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5240784-5240907 | 180.00 | -104.99 | 121 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5239239-5239363 | 175.00 | -99.88 | 123 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5240497-5240615 | 175.00 | -76.02 | 115 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5238847-5238950 | 163.00 | -92.08 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5238299-5238414 | 156.00 | -94.28 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582363 | chr18:5239376-5239484 | 146.00 | -86.86 | 102 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5240976-5241083 | 230.00 | -87.77 | 108 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5241328-5241444 | 225.00 | -95.55 | 122 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5239805-5239910 | 223.00 | -88.18 | 109 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5238599-5238708 | 218.00 | -93.01 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5239380-5239480 | 217.00 | -100.28 | 96 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5241584-5241693 | 211.00 | -98.76 | 117 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5240046-5240156 | 209.00 | -91.86 | 101 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5238155-5238263 | 205.00 | -103.78 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5238402-5238510 | 204.00 | -108.20 | 107 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5238985-5239103 | 203.00 | -97.28 | 119 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5241726-5241836 | 203.00 | -96.49 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5239156-5239259 | 201.00 | -86.24 | 110 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5240731-5240836 | 201.00 | -87.60 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5240839-5240942 | 198.00 | -96.04 | 110 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5240279-5240389 | 192.00 | -96.18 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5239905-5240023 | 191.00 | -92.97 | 117 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5238728-5238837 | 189.00 | -94.34 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5239476-5239593 | 189.00 | -86.49 | 118 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5240553-5240663 | 189.00 | -99.55 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5239272-5239395 | 180.00 | -104.99 | 121 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5241875-5241987 | 180.00 | -91.77 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5239670-5239801 | 170.00 | -90.85 | 129 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5238847-5238950 | 163.00 | -92.08 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5238299-5238414 | 156.00 | -94.28 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000582008 | chr18:5241087-5241202 | 154.00 | -91.63 | 116 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580082 | chr18:5238599-5238708 | 218.00 | -93.01 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580082 | chr18:5238155-5238263 | 205.00 | -103.78 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580082 | chr18:5238402-5238510 | 204.00 | -108.20 | 107 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580082 | chr18:5238728-5238837 | 189.00 | -94.34 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580082 | chr18:5238847-5238952 | 162.00 | -96.37 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580082 | chr18:5238299-5238414 | 156.00 | -94.28 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5238599-5238708 | 218.00 | -93.01 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5239457-5239557 | 217.00 | -100.28 | 96 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5239232-5239336 | 208.00 | -88.74 | 110 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5238985-5239103 | 203.00 | -97.28 | 119 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5238728-5238837 | 189.00 | -94.34 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5239349-5239472 | 180.00 | -104.99 | 121 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5239115-5239219 | 168.00 | -89.68 | 108 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000580684 | chr18:5238847-5238950 | 163.00 | -92.08 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5243259-5243366 | 230.00 | -87.77 | 108 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5243611-5243727 | 225.00 | -95.55 | 122 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5242088-5242193 | 223.00 | -88.18 | 109 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5241663-5241763 | 217.00 | -100.28 | 96 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5243867-5243976 | 211.00 | -98.76 | 117 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5242329-5242439 | 209.00 | -91.86 | 101 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5241438-5241542 | 208.00 | -88.74 | 110 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5244009-5244119 | 203.00 | -96.49 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5243014-5243119 | 201.00 | -87.60 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5243122-5243225 | 198.00 | -96.04 | 110 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5242562-5242672 | 192.00 | -96.18 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5242188-5242306 | 191.00 | -92.97 | 117 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5241759-5241876 | 189.00 | -86.49 | 118 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5242836-5242946 | 189.00 | -99.55 | 113 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5241109-5241221 | 188.00 | -104.54 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5240994-5241106 | 181.00 | -98.22 | 111 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5241555-5241678 | 180.00 | -104.99 | 121 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5244158-5244270 | 180.00 | -91.77 | 112 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5241953-5242084 | 170.00 | -90.85 | 129 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5241321-5241425 | 168.00 | -89.68 | 108 | NA | | ENSG00000263753 | hsa-mir-222 | ENST00000577439 | chr18:5243370-5243485 | 154.00 | -91.63 | 116 | NA |
 RNA A-to-I editing events in lncRNA. RNA A-to-I editing events in lncRNA. |
| LncRNAediting ID | LncRNA Ensembl ID | Chromosome | Editing Position | Strand | Gene Type | Gene Name | Transcript ID | Transcript Type | Transcript Name |
 Edited-associated DElncRNAs in cancer. Edited-associated DElncRNAs in cancer. |
| LncRNA Ensembl ID | LncRNA Index | Cancer Type | Chr_Postion_Strand | AVE1 | AVE2 | log2FC | W-value | P-value | Adjc.p-value | Change |
 Correlation between RNA A-to-I editing events's frequecy and lncRNA expression. Correlation between RNA A-to-I editing events's frequecy and lncRNA expression. |
| LncRNA Ensembl ID | LncRNA Index | Correlation | P-value | Adjc.p-value |
 Cis-expression quantitative trait loci(cis-eQTL) of lncRNA. Cis-expression quantitative trait loci(cis-eQTL) of lncRNA. |
| LncRNA Ensembl ID | LncRNA Name | SNP info | Number of Positive corelated Cancer | Positive corelated Cancer | Number of Negative corelated Cancer | Negative corelated Cancer |
 lncRNA regulates differentially expressed genes by function as enhancer. lncRNA regulates differentially expressed genes by function as enhancer. |
| LncRNA Ensembl ID | PC Gene ID | PC Gene Name | Positive correlated cancers | Cancer with PC gene up-regulation | Cancer with PC gene down-regulation |
 LncRNA-TF complex positively regulates the gene expression by target promoter region (#cancer types with positive correlation >= 5). LncRNA-TF complex positively regulates the gene expression by target promoter region (#cancer types with positive correlation >= 5). |
| lncRNA ID | TF ID | TF Name | PCgene ID | PCgene Name | Number of Cancer | Canaer Types |
 LncRNA-TF complex negatively regulates the gene expression by target promoter region (#cancer types with negative correlation >= 5). LncRNA-TF complex negatively regulates the gene expression by target promoter region (#cancer types with negative correlation >= 5). |
| lncRNA ID | TF ID | TF Name | PCgene ID | PCgene Name | Number of Cancer | Canaer Types |
 LncRNA-RBP complex positively regulates the exon skippping events by target skipped eoxon region. LncRNA-RBP complex positively regulates the exon skippping events by target skipped eoxon region. |
| LncRNA Ensembl ID | RBP ID | RBP Gene Name | Exon Skipping ID | Skipped Exon | EX Gene Name | EX Affected TransID | ORF_anno | Cancer Type |
 LncRNA-RBP complex negatively regulates the exon skippping events by target skipped eoxon region. LncRNA-RBP complex negatively regulates the exon skippping events by target skipped eoxon region. |
| LncRNA Ensembl ID | RBP ID | RBP Gene Name | Exon Skipping ID | Skipped Exon | EX Gene Name | EX Affected TransID | ORF_anno | Cancer Type |
 lncRNA regulates differential expressed mRNA by directly targeting 3' UTR region. lncRNA regulates differential expressed mRNA by directly targeting 3' UTR region. |
| LncRNA Ensembl ID | LncRNA ENST ID | PC Gene Name | PC Gene ID | PC ENST ID | dG | nDG | Cancer with PC gene up-regulation | Cancer with PC gene Down-regulation |
 lncRNA regulates mRNA by competing the miRNA binding site with mRNA. lncRNA regulates mRNA by competing the miRNA binding site with mRNA. |
| LncRNA Ensembl ID | lncRNA-miRNA-mRNA | Cancer Types | | ENSG00000263753 | LINC00667,hsa-mir-221,ZBED3 | PCPG,KICH,THCA,LGG,GBM | | ENSG00000263753 | LINC00667,hsa-mir-22,WDR33 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,ZNF22 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,CRMP1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PHF2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,ARL6IP6 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,CERS5 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,COIL | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,YTHDC1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PRR14 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,C2orf68 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,NRF1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PCGF2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,ZNF7 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,NAA16 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,UNK | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,RCE1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,RFXAP | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,ZSWIM4 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,EHMT2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PHC1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,EPC1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,ZCCHC8 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PAXBP1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PAGR1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,IRF2BP2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,MBTD1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,TIA1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,DPF2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,KPNA5 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,EDRF1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,SBK1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,RBMX | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,H3-3B | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,FBXO46 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,SET | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,SUMO2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,RGL2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,RBM14 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PCIF1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,HMGB1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,DGCR8 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,ENDOV | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,FANCG | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,CTDSPL2 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,RBM22 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,WRNIP1 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,PRR3 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,SCAF4 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,KDM5B | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-22,TTC31 | PCPG,KICH,CHOL,LGG,KIRP | | ENSG00000263753 | LINC00667,hsa-mir-222,MARCHF8 | KICH,TGCT,THCA,LGG,GBM | | ENSG00000263753 | LINC00667,hsa-mir-222,LCMT2 | PCPG,KICH,TGCT,LGG,GBM | | ENSG00000263753 | LINC00667,hsa-mir-222,IKZF5 | PCPG,KICH,TGCT,LGG,GBM | | ENSG00000263753 | LINC00667,hsa-mir-222,DHTKD1 | KICH,TGCT,THCA,LGG,GBM | | ENSG00000263753 | LINC00667,hsa-mir-222,ZSWIM8 | PCPG,KICH,TGCT,LGG,GBM |
 ORFfinder result for the gencode.v22.lncRNA.transcript.fa. ORFfinder result for the gencode.v22.lncRNA.transcript.fa. |
| lncRNA Ensembl ID | lncRNA ENST ID | length(AA) | start at transcript | end at transcript | | ENSG00000263753.5 | ENST00000581471.2 | 165 | 655 | 158 | | ENSG00000263753.5 | ENST00000582363.1 | 165 | 117 | 614 | | ENSG00000263753.5 | ENST00000582008.4 | 77 | 612 | 845 | | ENSG00000263753.5 | ENST00000580082.1 | 126 | 3541 | 3921 | | ENSG00000263753.5 | ENST00000580684.4 | 88 | 568 | 831 | | ENSG00000263753.5 | ENST00000577439.1 | 77 | 146 | 379 |
|

 Gene summary
Gene summary AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure.
AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure.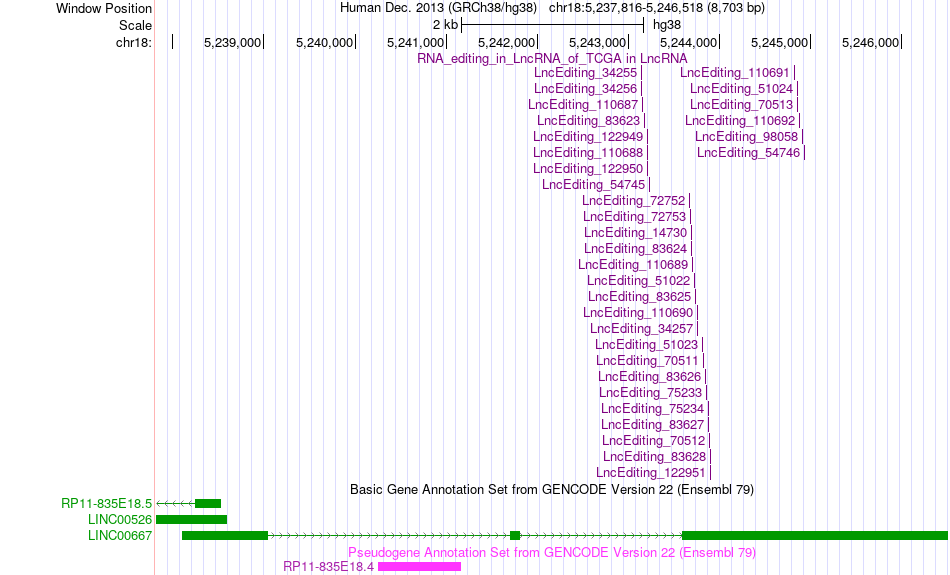
 Differentially expressed gene analysis between cancer and normal samples.
Differentially expressed gene analysis between cancer and normal samples. Correlation between lncRNAs and cancer stages.
Correlation between lncRNAs and cancer stages.
 lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5).
lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5). lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5).
lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5). lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5).
lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5).







