
|
||||||
|
lncRNA: ENSG00000262879 |
Summary for RP11-156P1.3 |
 Gene summary Gene summary |
| Gene information | Ensembl ID | ENSG00000262879 | Gene symbol | RP11-156P1.3 |
| Gene name | ||
| HGNC | ||
| Entrez ID | 101927060 | |
| Gene type | processed_transcript | |
| Synonyms | ||
| UniProtAcc |
Top |
Structure and expression level for RP11-156P1.3 |
 AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure. AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure. |
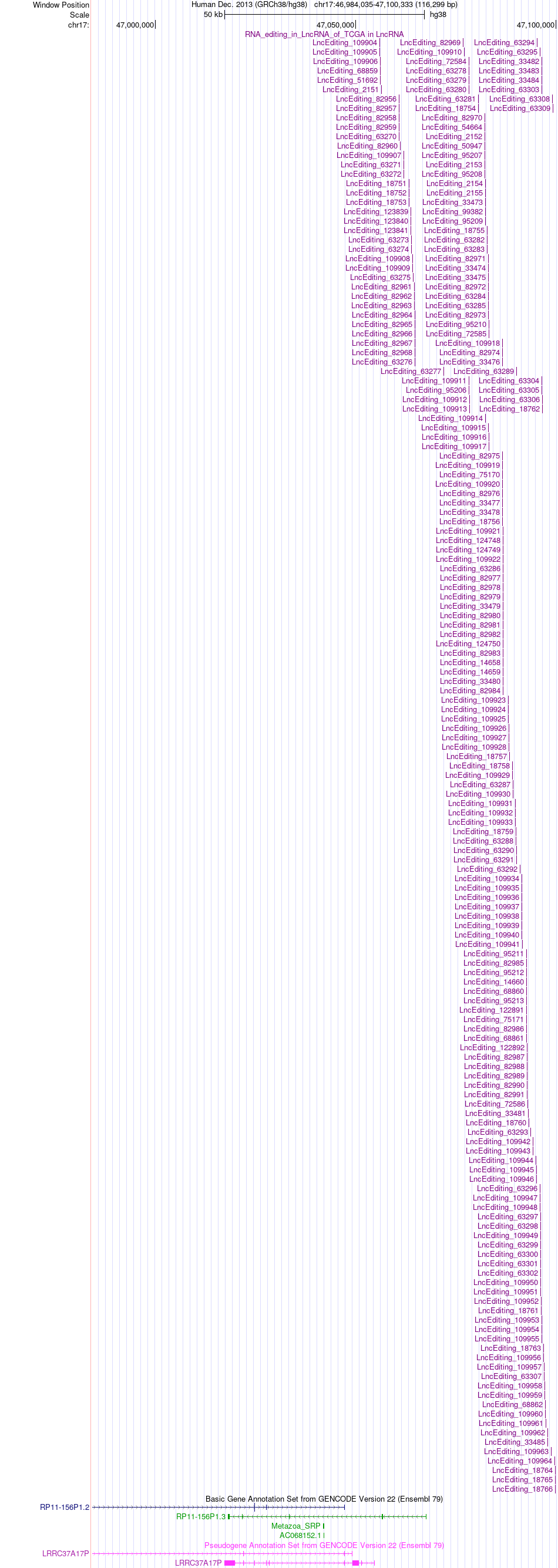 |
 Differentially expressed gene analysis between cancer and normal samples. Differentially expressed gene analysis between cancer and normal samples. |
| Ensembl ID | Base mean | log2FC(AD/control) | lfcSE | Stat | P-value | Adjc.p-value | DEG direction | Cancer type |
 Correlation between lncRNAs and cancer stages. Correlation between lncRNAs and cancer stages. |
| LncRNA Ensembl ID | Correlation | P-value | Cancer Type | Adjc.p-value |
| ENSG00000262879 | -4.613812e-01 | 5.349699e-06 | KICH | 1.224960e-05 |
Top |
lncRNA-Protein interaction for RP11-156P1.3 |
 |
 lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5). lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5). |
| LncRNA Ensembl ID | LncRNA ENST ID | LncRNA Name | Protein Ensembl ID | Protein Name | CatRAPID Score | Lncpro Score | UniPortID | Gene Subtype | Public source |
| ENSG00000262879 | ENST00000574741 | RP11-156P1.3 | ENSG00000227345 | PARG | 66.87 | 92.5939 | Q86W56 | Epi | NA |
| ENSG00000262879 | ENST00000574741 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 52.49 | 90.3043 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000574741 | RP11-156P1.3 | ENSG00000164190 | NIPBL | 50.66 | 92.9772 | Q6KC79 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000067369 | TP53BP1 | 56.33 | 90.9959 | Q12888 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000128908 | INO80 | 52.54 | 92.6051 | Q9ULG1 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000149311 | ATM | 51.11 | 94.2089 | Q13315 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000024048 | UBR2 | 47.94 | 92.7453 | Q8IWV8 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 47.6 | 91.837 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 46.87 | 91.1062 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000173473 | SMARCC1 | 46.46 | 91.5964 | Q92922 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000175054 | ATR | 43.61 | 92.4938 | Q13535 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000196367 | TRRAP | 43.3 | 93.3716 | Q9Y4A5 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000253729 | PRKDC | 42.64 | 94.9984 | P78527 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000085224 | ATRX | 41.08 | 91.7642 | P46100 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000085224 | ATRX | 40.93 | 91.4368 | P46100 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000146247 | PHIP | 37.98 | 93.6448 | Q8WWQ0 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000141027 | NCOR1 | 36.57 | 93.2562 | O75376 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 36.3 | 95.348 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000147130 | ZMYM3 | 36.23 | 92.2849 | Q14202 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000183337 | BCOR | 34.94 | 90.0316 | Q6W2J9 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000085224 | ATRX | 32.97 | 91.2216 | P46100 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000185658 | BRWD1 | 32.57 | 93.8937 | Q9NSI6 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000139613 | SMARCC2 | 31.74 | 92.6021 | Q8TAQ2 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000124177 | CHD6 | 30.94 | 91.2729 | Q8TD26 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000141027 | NCOR1 | 30.36 | 90.7208 | O75376 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000167548 | KMT2D | 29.24 | 92.5384 | O14686 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000198646 | NCOA6 | 27.55 | 92.5452 | Q14686 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000171634 | BPTF | 27.07 | 94.8582 | Q12830 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000163935 | SFMBT1 | 26.06 | 90.9787 | Q9UHJ3 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000167548 | KMT2D | 24.41 | 94.3182 | O14686 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000067369 | TP53BP1 | 22.36 | 91.6606 | Q12888 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000197323 | TRIM33 | 20.35 | 90.3784 | Q9UPN9 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 18.13 | 92.5348 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000156650 | KAT6B | 17.97 | 94.5778 | Q8WYB5 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000085224 | ATRX | 16.73 | 93.9837 | P46100 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000141027 | NCOR1 | 15.21 | 92.0019 | O75376 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000171634 | BPTF | 12.56 | 90.2627 | Q12830 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000140396 | NCOA2 | 12.41 | 90.054 | Q15596 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000049618 | ARID1B | 8.3 | 93.1676 | Q8NFD5 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000128908 | INO80 | 64.27 | 91.1266 | Q9ULG1 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000141027 | NCOR1 | 45.84 | 90.6191 | O75376 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000139613 | SMARCC2 | 40.4 | 90.903 | Q8TAQ2 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000167548 | KMT2D | 36.71 | 91.7648 | O14686 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000156650 | KAT6B | 24.43 | 92.2635 | Q8WYB5 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000123636 | BAZ2B | 55.45 | 93.5011 | Q9UIF8 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000025293 | PHF20 | 54.38 | 92.2462 | Q9BVI0 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571901 | RP11-156P1.3 | ENSG00000048649 | RSF1 | 52.76 | 92.7803 | Q96T23 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000054267 | ARID4B | 48.84 | 91.0487 | Q4LE39 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000009954 | BAZ1B | 47.27 | 94.2659 | Q9UIG0 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000100888 | CHD8 | 45.02 | 90.4203 | Q9HCK8 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000005483 | KMT2E | 44.71 | 91.472 | Q8IZD2 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000005483 | KMT2E | 44.43 | 90.8861 | Q8IZD2 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000181555 | SETD2 | 44.01 | 91.9763 | Q9BYW2 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000165671 | NSD1 | 43.03 | 90.8653 | Q96L73 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000165671 | NSD1 | 43.03 | 90.8394 | Q96L73 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000181555 | SETD2 | 42.9 | 94.3777 | Q9BYW2 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000163939 | PBRM1 | 41.77 | 91.7867 | Q86U86 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000117139 | KDM5B | 41.39 | 92.2608 | Q9UGL1 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000108557 | RAI1 | 40.75 | 91.9904 | Q7Z5J4 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000055609 | KMT2C | 39.96 | 92.3379 | Q8NEZ4 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000005339 | CREBBP | 39.7 | 90.5711 | Q92793 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000055609 | KMT2C | 39.58 | 92.8025 | Q8NEZ4 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000073614 | KDM5A | 39.13 | 92.1567 | P29375 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000086758 | HUWE1 | 39.09 | 91.204 | Q7Z6Z7 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000086758 | HUWE1 | 38.93 | 95.5194 | Q7Z6Z7 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000129292 | PHF20L1 | 38.79 | 90.6222 | A8MW92 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000118900 | UBN1 | 38.68 | 91.5876 | Q9NPG3 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000032219 | ARID4A | 38.6 | 91.3999 | P29374 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000092201 | SUPT16H | 37.95 | 90.7478 | Q9Y5B9 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000198919 | DZIP3 | 36.85 | 90.8388 | Q86Y13 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000170322 | NFRKB | 36.2 | 90.6384 | Q6P4R8 | Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000171988 | JMJD1C | 35.36 | 92.3962 | Q15652 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000164190 | NIPBL | 35.09 | 93.9362 | Q6KC79 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000164190 | NIPBL | 35.04 | 90.0947 | Q6KC79 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000171988 | JMJD1C | 34.8 | 93.4126 | Q15652 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000165671 | NSD1 | 34.36 | 92.4153 | Q96L73 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000078403 | MLLT10 | 34.26 | 90.8163 | P55197 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 34.2 | 93.3275 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000168769 | TET2 | 33.41 | 90.768 | Q6N021 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000005339 | CREBBP | 32.74 | 93.3833 | Q92793 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000165288 | BRWD3 | 32.6 | 94.1853 | Q6RI45 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000100393 | EP300 | 31.98 | 93.5095 | Q09472 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000055609 | KMT2C | 30.44 | 94.2981 | Q8NEZ4 | Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000137337 | MDC1 | 28.81 | 93.0883 | Q14676 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000183495 | EP400 | 28.11 | 92.1181 | Q96L91 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000076108 | BAZ2A | 27.95 | 90.7002 | Q9UIF9 | TF,RBP,Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000126012 | KDM5C | 26.44 | 91.8082 | P41229 | Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000168769 | TET2 | 26.25 | 92.966 | Q6N021 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000083168 | KAT6A | 25.93 | 93.1919 | Q92794 | Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000083168 | KAT6A | 25.29 | 91.959 | Q92794 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000025293 | PHF20 | 22.76 | 90.4182 | Q9BVI0 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000083168 | KAT6A | 21.61 | 91.3184 | Q92794 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000048649 | RSF1 | 19.8 | 90.7111 | Q96T23 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000054267 | ARID4B | 19.25 | 90.3926 | Q4LE39 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000009954 | BAZ1B | 19.05 | 91.6482 | Q9UIG0 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000171681 | ATF7IP | 18.92 | 91.1283 | Q6VMQ6 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000181555 | SETD2 | 18.39 | 92.979 | Q9BYW2 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000227345 | PARG | 17.82 | 90.133 | Q86W56 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000165671 | NSD1 | 17.79 | 92.2428 | Q96L73 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000005483 | KMT2E | 17.73 | 90.2803 | Q8IZD2 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000108557 | RAI1 | 17.06 | 91.7908 | Q7Z5J4 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000129292 | PHF20L1 | 16.55 | 90.3458 | A8MW92 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000005339 | CREBBP | 16.42 | 90.4237 | Q92793 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000055609 | KMT2C | 15.09 | 90.1049 | Q8NEZ4 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000164190 | NIPBL | 14.37 | 94.2468 | Q6KC79 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 13.98 | 90.2267 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000168769 | TET2 | 13.4 | 90.126 | Q6N021 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000137337 | MDC1 | 11.4 | 91.6552 | Q14676 | Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000117713 | ARID1A | 8.52 | 92.8931 | O14497 | Epi | NA |
| ENSG00000262879 | ENST00000574741 | RP11-156P1.3 | ENSG00000196199 | MPHOSPH8 | 64.79 | 92.2108 | Q99549 | Epi | NA |
| ENSG00000262879 | ENST00000574741 | RP11-156P1.3 | ENSG00000139613 | SMARCC2 | 48.97 | 94.6925 | Q8TAQ2 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000025293 | PHF20 | 68.4 | 90.372 | Q9BVI0 | TF,Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000009954 | BAZ1B | 57.57 | 90.9211 | Q9UIG0 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000164190 | NIPBL | 43.85 | 92.4542 | Q6KC79 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000171988 | JMJD1C | 43.29 | 91.3146 | Q15652 | Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 42.41 | 91.732 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000571665 | RP11-156P1.3 | ENSG00000078403 | MLLT10 | -9.62 | 90.4852 | P55197 | Epi | NA |
 lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5). lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5). |
| LncRNA Ensembl ID | LncRNA ENST ID | LncRNA Name | Protein Ensembl ID | Protein Name | CatRAPID Score | Lncpro Score | UniPortID | Gene Subtype | Public source |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000102189 | EEA1 | 57.52 | 90.6224 | Q15075 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000123636 | BAZ2B | 56.48 | 93.4474 | Q9UIF8 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000123636 | BAZ2B | 55.45 | 93.9657 | Q9UIF8 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000025293 | PHF20 | 54.42 | 90.4809 | Q9BVI0 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000025293 | PHF20 | 54.38 | 92.8054 | Q9BVI0 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000096401 | CDC5L | 49.17 | 90.5368 | Q99459 | TF,RBP | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000144747 | TMF1 | 48.52 | 92.083 | P82094 | TF | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000144747 | TMF1 | 47.99 | 90.7177 | P82094 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 47.6 | 95.0741 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 46.87 | 91.299 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000102189 | EEA1 | 44.22 | 91.3344 | Q15075 | TF | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000117139 | KDM5B | 41.39 | 91.7586 | Q9UGL1 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000100207 | TCF20 | 41.14 | 91.1062 | Q9UGU0 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000100207 | TCF20 | 40.98 | 93.0598 | Q9UGU0 | TF | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000115816 | CEBPZ | 40.47 | 91.6937 | Q03701 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000116539 | ASH1L | 38.11 | 92.0791 | Q9NR48 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 36.3 | 95.4892 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000187605 | TET3 | 35.77 | 90.3399 | O43151 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000168769 | TET2 | 33.41 | 93.1974 | Q6N021 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000100207 | TCF20 | 32.55 | 92.0528 | Q9UGU0 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000171634 | BPTF | 32.39 | 90.8796 | Q12830 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000084093 | REST | 31.26 | 91.3547 | Q13127 | TF,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000084093 | REST | 30.93 | 90.2349 | Q13127 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000167232 | ZNF91 | 29.98 | 92.1653 | Q05481 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000198521 | ZNF43 | 28.66 | 90.0553 | P17038 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000117000 | RLF | 28.01 | 92.2397 | Q13129 | TF | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000076108 | BAZ2A | 27.95 | 91.0303 | Q9UIF9 | TF,RBP,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000171634 | BPTF | 27.07 | 94.9764 | Q12830 | TF,Epi | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000168769 | TET2 | 26.25 | 93.5285 | Q6N021 | TF,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000125812 | GZF1 | 24.26 | 90.5516 | Q9H116 | TF | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000169554 | ZEB2 | 24.13 | 90.5779 | O60315 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000140836 | ZFHX3 | 23.91 | 93.7225 | Q15911 | TF | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000140836 | ZFHX3 | 23.04 | 92.0343 | Q15911 | TF | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000025293 | PHF20 | 22.76 | 90.9133 | Q9BVI0 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000102189 | EEA1 | 22.47 | 91.0483 | Q15075 | TF | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000117000 | RLF | 22.41 | 93.9504 | Q13129 | TF | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000106571 | GLI3 | 21.37 | 93.1796 | P10071 | TF | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000140836 | ZFHX3 | 19.7 | 92.5611 | Q15911 | TF | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000118058 | KMT2A | 18.13 | 92.6803 | Q03164 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000100207 | TCF20 | 16.75 | 92.9661 | Q9UGU0 | TF | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000168769 | TET2 | 13.4 | 90.3401 | Q6N021 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000171634 | BPTF | 12.56 | 90.4777 | Q12830 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000140396 | NCOA2 | 12.41 | 90.3061 | Q15596 | TF,Epi | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000140836 | ZFHX3 | 8.89 | 91.6791 | Q15911 | TF | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000105750 | ZNF85 | 6.31 | 90.1065 | Q03923 | TF | NA |
| ENSG00000262879 | ENST00000574741 | RP11-156P1.3 | ENSG00000256223 | ZNF10 | 22.08 | 90.3686 | P21506 | TF | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000025293 | PHF20 | 68.4 | 91.4021 | Q9BVI0 | TF,Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000174282 | ZBTB4 | 68.35 | 90.0306 | Q9P1Z0 | TF | NA |
 lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5). lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5). |
| LncRNA Ensembl ID | LncRNA ENST ID | LncRNA Name | Protein Ensembl ID | Protein Name | CatRAPID Score | Lncpro Score | UniPortID | Gene Subtype | Public source |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 42.41 | 90.994 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000570314 | RP11-156P1.3 | ENSG00000167978 | SRRM2 | 14.72 | 91.8089 | Q9UQ35 | RBP | NA |
| ENSG00000262879 | ENST00000571665 | RP11-156P1.3 | ENSG00000196943 | NOP9 | 1.14 | 90.3045 | Q86U38 | RBP | NA |
| ENSG00000262879 | ENST00000574741 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 52.49 | 90.5014 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000056097 | ZFR | 56.08 | 90.363 | Q96KR1 | RBP | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000056097 | ZFR | 55.76 | 90.512 | Q96KR1 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000159082 | SYNJ1 | 44.21 | 91.0904 | O43426 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000129566 | TEP1 | 40.15 | 93.4289 | Q99973 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000114127 | XRN1 | 39.69 | 92.6883 | Q8IZH2 | RBP | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000198000 | NOL8 | 38.65 | 90.7975 | Q76FK4 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000165934 | CPSF2 | 36.99 | 90.9397 | Q9P2I0 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000198919 | DZIP3 | 36.85 | 91.0257 | Q86Y13 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000091009 | RBM27 | 36.07 | 90.2698 | Q9P2N5 | RBP | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000137776 | SLTM | 36.05 | 93.3357 | Q9NWH9 | RBP | ENCODE |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000139746 | RBM26 | 35.06 | 90.4676 | Q5T8P6 | RBP | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 34.2 | 93.7119 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000021776 | AQR | 33.72 | 92.7372 | O60306 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000125676 | THOC2 | 33.37 | 92.84 | Q8NI27 | RBP | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000162775 | RBM15 | 32.92 | 91.3694 | Q96T37 | RBP | ENCODE |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000198000 | NOL8 | 30.98 | 91.0968 | Q76FK4 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000067596 | DHX8 | 30.73 | 90.401 | Q14562 | RBP | NA |
| ENSG00000262879 | ENST00000575173 | RP11-156P1.3 | ENSG00000129317 | PUS7L | 30.27 | 90.6958 | Q9H0K6 | RBP | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000076108 | BAZ2A | 27.95 | 91.2281 | Q9UIF9 | TF,RBP,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000133226 | SRRM1 | 16.9 | 92.9447 | Q8IYB3 | RBP | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000133226 | SRRM1 | 15.78 | 93.5966 | Q8IYB3 | RBP | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000133226 | SRRM1 | 14.06 | 91.9629 | Q8IYB3 | RBP | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000012048 | BRCA1 | 13.98 | 90.3957 | P38398 | RBP,Epi | NA |
| ENSG00000262879 | ENST00000575930 | RP11-156P1.3 | ENSG00000167978 | SRRM2 | 12.56 | 90.5886 | Q9UQ35 | RBP | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000167978 | SRRM2 | 12.16 | 96.9515 | Q9UQ35 | RBP | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000167978 | SRRM2 | 9.86 | 94.9754 | Q9UQ35 | RBP | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000111605 | CPSF6 | 7.96 | 92.0243 | Q16630 | RBP | NA |
| ENSG00000262879 | ENST00000572864 | RP11-156P1.3 | ENSG00000100461 | RBM23 | 5.1 | 92.741 | Q86U06 | RBP | NA |
| ENSG00000262879 | ENST00000576938 | RP11-156P1.3 | ENSG00000167978 | SRRM2 | 4.42 | 94.0234 | Q9UQ35 | RBP | NA |
| ENSG00000262879 | ENST00000571143 | RP11-156P1.3 | ENSG00000149532 | CPSF7 | 3.94 | 90.586 | Q8N684 | RBP | High-throughput data |
Top |
|








