
|
||||||
|
lncRNA: ENSG00000261068 |
Summary for RP11-7K24.3 |
 Gene summary Gene summary |
| Gene information | Ensembl ID | ENSG00000261068 | Gene symbol | RP11-7K24.3 |
| Gene name | ||
| HGNC | ||
| Entrez ID | ||
| Gene type | lincRNA | |
| Synonyms | ||
| UniProtAcc |
Top |
Structure and expression level for RP11-7K24.3 |
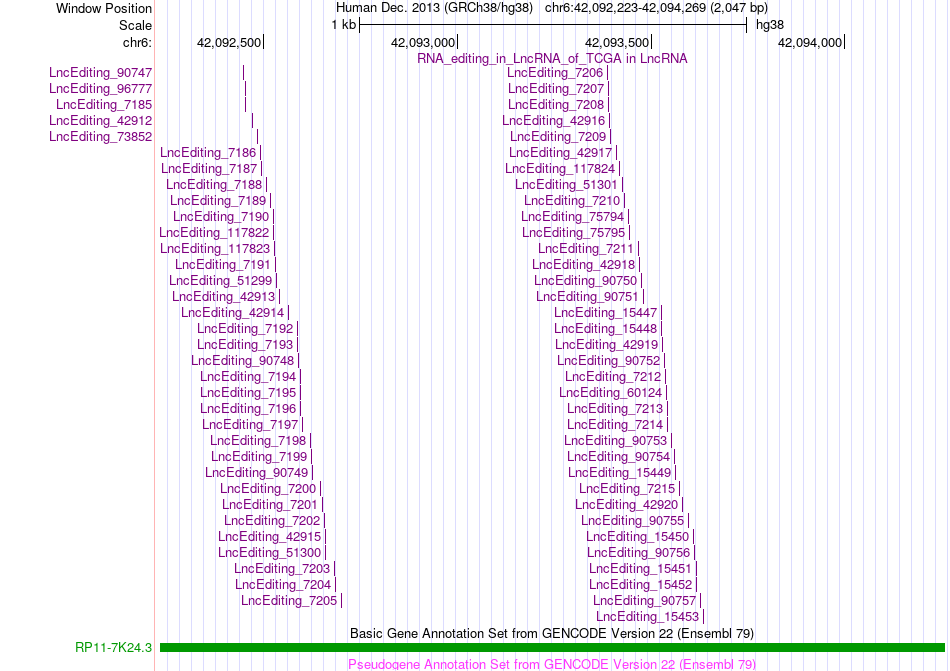
 AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure. AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure. |
 |
 Differentially expressed gene analysis between cancer and normal samples. Differentially expressed gene analysis between cancer and normal samples. |
| Ensembl ID | Base mean | log2FC(AD/control) | lfcSE | Stat | P-value | Adjc.p-value | DEG direction | Cancer type |
| ENSG00000261068 | 1.154483e+02 | 5.484813e+00 | 5.914667e-01 | 9.273241e+00 | 1.810000e-20 | 9.450000e-19 | UP | CHOL |
| ENSG00000261068 | 5.389577e+02 | 3.103097e+00 | 8.081337e-01 | 3.839831e+00 | 1.231190e-04 | 1.441792e-03 | UP | CESC |
| ENSG00000261068 | 8.064491e+02 | -2.141447e+00 | 2.155082e-01 | -9.936732e+00 | 2.880000e-23 | 8.340000e-22 | DOWN | HNSC |
| ENSG00000261068 | 1.981268e+02 | 1.537333e+00 | 3.012583e-01 | 5.103040e+00 | 3.340000e-07 | 1.570000e-06 | UP | KIRP |
| ENSG00000261068 | 1.442076e+02 | 1.168797e+00 | 2.384019e-01 | 4.902634e+00 | 9.460000e-07 | 3.980000e-06 | UP | UCEC |
| ENSG00000261068 | 8.188087e+01 | 1.027507e+00 | 1.936946e-01 | 5.304777e+00 | 1.130000e-07 | 4.810000e-07 | UP | COAD |
| ENSG00000261068 | 4.412607e+02 | -2.058241e+00 | 2.920356e-01 | -7.047912e+00 | 1.820000e-12 | 3.460000e-11 | DOWN | STAD |
 Correlation between lncRNAs and cancer stages. Correlation between lncRNAs and cancer stages. |
| LncRNA Ensembl ID | Correlation | P-value | Cancer Type | Adjc.p-value |
| ENSG00000261068 | 4.869245e-01 | 6.939820e-04 | CHOL | 1.279910e-03 |
Top |
lncRNA-Protein interaction for RP11-7K24.3 |
 |
 lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5). lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5). |
| LncRNA Ensembl ID | LncRNA ENST ID | LncRNA Name | Protein Ensembl ID | Protein Name | CatRAPID Score | Lncpro Score | UniPortID | Gene Subtype | Public source |
 lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5). lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5). |
| LncRNA Ensembl ID | LncRNA ENST ID | LncRNA Name | Protein Ensembl ID | Protein Name | CatRAPID Score | Lncpro Score | UniPortID | Gene Subtype | Public source |
| ENSG00000261068 | ENST00000562471 | RP11-7K24.3 | ENSG00000188786 | MTF1 | 53.59 | 92.6687 | Q14872 | TF | NA |
 lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5). lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5). |
| LncRNA Ensembl ID | LncRNA ENST ID | LncRNA Name | Protein Ensembl ID | Protein Name | CatRAPID Score | Lncpro Score | UniPortID | Gene Subtype | Public source |
Top |
|








