| SPICE1,NEK4,CMTM4,SCAPER,KLHL42,ZFP28,ZFP14,HEATR1,AP5M1,RNF123,RBBP6,TBC1D5,MAP4K3,BECN1,RDX,TCF20,TAF4,PPP1R12A,GNL3L,TMEM161B,KIAA1109,STAU2,KAT6B,C11orf58,SMG1,CREBBP,PBRM1,SRRM2,TRAK1,SCRN1,ANKRD28,UBAP2,VPS13B,SACM1L,FGD4,GMPS,L2HGDH,TDG,NECAP1,PRRC2C,ATP11B,DNM1L,SNX19,PDPK1,R3HDM2,PHF3,ACBD5,CPEB4,LONP2,ARID1B,STRN,NMD3,TMEM33,TTC3,PHF20,KIDINS220,CDC27,REPS1,GTF2H3,RNF31,CLASP2,FBXO11,DDX6,GANAB,SEC23A,MKLN1,ARHGEF9,ITSN2,RNF38,RB1CC1,KLF13,SMAD1,KIF21A,MYCBP2,AGFG1,SNRNP200,MKRN2,MED12,POC1B,MSH6,FOXN3,GTF3C3,TRA2B,RNF138,OSBPL8,CPSF7,NCOA4,AZI2,TMOD2,ACTR2,DCAF7,EEF2,GPATCH8,ALMS1,RPAP1,ACTR8,MED1,NUP58,HBP1,EIF4G2,CDC23,NIN,NAA50,CUL2,GON4L,HUWE1,SLC25A40,YME1L1,ZZEF1,TLK1,RABEP1,FAM13B,MED23,DYNC2I1,EPM2AIP1,ICE2,DPP8,BCKDHB,RPRD1A,FYCO1,SPOPL,DOP1A,NOMO2,LRP6,FAM168B,PHF6,FAM53C,USP4,SINHCAF,HACD3,PFKM,REV1,FRS2,PNN,MLXIP,EDC4,UBXN7,PTPN23,RNF214,TAF4B,DCTN4,GPATCH2L,ARIH1,PUM1,SEC24B,FOXN2,LIMD1,ZFP1,KMT2D,CTNNB1,FUBP1,HUS1,ANAPC1,MON2,CEP57,RSBN1,RNF111,TP53BP1,TUBGCP4,WDR48,STRBP,PPIP5K2,G3BP1,GIGYF2,NCBP1,TFCP2,WBP11,TTC28,TMPO,DNMT1,SRP54,BRD2,PAN3,AFF4,DDX3X,ALG10B,IPO5,ASXL2,PHF8,RAF1,POLR2B,ESF1,BAZ2A,CEP350,PIKFYVE,R3HDM1,RAB3GAP2,DDHD2,CHD7,ASH1L,RAB18,DENND4A,GNPTAB,USP19,METTL6,RAP1B,WDR7,TMEM131,EP400,GNE,RSBN1L,SMG6,ADNP2,WDR20,NUP98,KIAA0586,TRIM23,CDK12,AKT3,GTF2A1,CHD9,TFIP11,MPHOSPH8,SOS1,SMAD5,CSNK1A1,TUBGCP3,TTC37,TNPO2,ELF1,CLINT1,PIGO,RPE,NSD2,TAF2,GCC2,CHD6,NEU3,DOCK10,PLAA,TRIM33,CPSF6,MED21,HEATR5A,DMTF1,FEM1B,HDAC4,THOC2,TEX10,CELF1,CCNK,NEMF,CTDSP2,DDX55,SDAD1,TAF5L,SCAF11,CNOT1,NF1,DIABLO,TOPBP1,MAPK8,CEBPZ,SLC11A2,NCBP3,RBMX,SECISBP2L,UBR1,NOP9,CHD8,TAOK3,TSR1,ILF3,RBM28,ARHGAP21,SCAI,DCUN1D1,RAB5A,ERBIN,PCBP1,INTS3,ZC3H11A,VGLL4,RIC1,MARF1,KTN1,C5orf24,PARP2,PTPN4,FARP2,ZDHHC17,CLIP1,SART3,RAD50,API5,NCOA2,RC3H2,XPO1,PRR14L,RNF169,UBR5,KLHL24,ACACA,USP42,PRKAR2A,MIER3,USP9X,ROCK2,CCDC88A,SATB1,MTMR1,SEC23IP,WNK1,ASXL1,PPP4R2,SOCS7,PFAS,RBM33,RNF44,SMCHD1,LYSMD3,PSIP1,CCNG1,PPARA,GFM2,USP7,KIF2A,REST,EIF3L,EMSY,G2E3,SOS2,LARS2,CS,POLK,RANBP2,RTF1,DYRK1A,AFF1,BLOC1S6,MECP2,CARM1,PICALM,USP14,RAD54L2,CKAP5,CPSF2,SPG11,FAM172A,ATP2B1,GLYR1,DHX36,MBTD1,ZNF91,RALGAPA1,FER,RAB6A,GSK3B,AEBP2,CEP170,OXSR1,NT5C2,ZMYND11,RALGAPB,HIPK1,QTRT2,NSD3,ZMYM5,PTPN11,WDR36,MCM3AP,ZNF84,PLEKHB2,ZNF17,TSC1,NR2C2,DIP2A,CLTC,ATRN,AP1AR,SLC25A36,PARG,EPB41L5,ATP2A2,SON,PTPRG,M6PR,APAF1,BBX,TAF1,THAP9,PCNX1,DMXL2,DCAF16,EIF2A,ZNF720,LYRM7,TTBK2,SMPD4,SLMAP,IRAK1BP1,RBL1,CIPC,STRAP,ARHGAP5,BCLAF1,FTO,SEC14L1,SLC7A6,SETD5,SF3B1,NIPA2,C2CD5,PCM1,DYNC1LI2,KBTBD4,ZNF106,CERT1,UBXN4,SENP6,MED13L,SF3A1,SIK3,KMT2B,UBE3A,NCOA3,SYNJ1,SBF2,TNPO1,SPTLC1,FAM160B1,RO60,NCAPD3,HEATR5B,BIRC6,SEC31A,TMEM184C,ARL5A,DPY19L3,BDP1,BCOR,SYNRG,SCAF4,EIF2AK4,PDS5A,USP10,TMEM131L,AFAP1,NDUFS1,NSD1,MPHOSPH9 |

 Gene summary
Gene summary AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure.
AS events and RNA A-to-I editing events of lncRNA in TCGA based on Genvode V22 structure.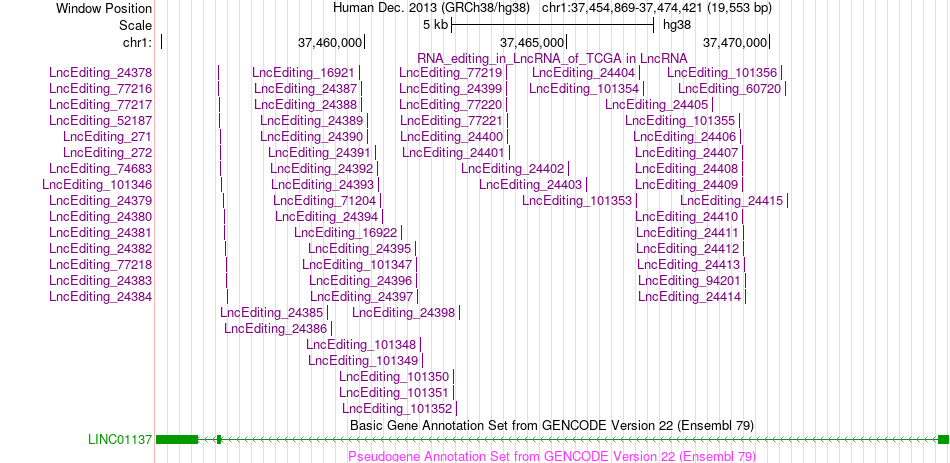
 Differentially expressed gene analysis between cancer and normal samples.
Differentially expressed gene analysis between cancer and normal samples. Correlation between lncRNAs and cancer stages.
Correlation between lncRNAs and cancer stages.
 lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5).
lncRNA-epigenetic factor interaction (#cancer types with positive correlation >= 5). lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5).
lncRNA-transcript factor interaction (#cancer types with positive correlation >= 5). lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5).
lncRNA-RNA binding protein interaction (#cancer types with positive correlation >= 5).







