
|
Home |
Download |
Statistics |
Examples |
Help |
Contact |
About SexAnnoDB
Sex differences are a growing focus of cancer incidence and mortality studies. Recently, personalized approaches in cancer treatment have taken sex differences into account. However, there are no systematic and intensive analyses of different sex groups until now. Here we built SexAnnoDB(A comprehensive sex annotation database in cancer) to provide a resource and reference for intensive functional annotations of sex effects in cancer to facilitate the development of personalized precision medicine. In this database, we analyzed different levels of molecules, including molecules of genome, epigenome, transcriptome, and proteome of TCGA across 27 cancer types. For each molecule, we performed multiple systematic and bioinformatic analyses, including male-specific differential molecules, female-specific differential molecules, and sex-biased molecules. What's more, we analyzed transcript factor-gene regulation network, RNA binding protein-exon skipping regulation network, ceRNA of female and male groups, diverse variant information (Sex-biased Expression Quantitative Trait Loci (eQTL), Sex-biased Splicing Quantitative Trait Loci (sQTL)), and methylation information (Sex-biased Expression Quantitative Trait Methylation(eQTM), Sex-biased Splicing Quantitative Trait Methylation (sQTM)). SexAnnoDB is the first database that systematically and intensively annotates the sex effect across differential biomolecules in cancer, which will be a unique resource for cancer and drug research communities to develop personalized precision medicine plans for patients.
 Search by gene
Search by gene|
∗Through search by gene, users are able to access the general information and sex-biased regulations of genes. |
 Search by drug
Search by drug
|
∗SexAnnoDB provided 205 cancer therapeutic target genes that were targeted by 206 drugs for 40 cancer types from the National Cancer Institute. Through search by drug, users are able to access the drug target genes and the general information and sex-biased regulations of genes. |
 Summary of sex-related regulation for each cancer type
Summary of sex-related regulation for each cancer type∗Through the browser for each cancer type, users are able to get the summary of cancer, the top 20 sex-biased TF-Gene regulations,top20 sex-biased RBP-ES regulations, sex-biased cis-eQTL regulations, sex-biased cis-sQTL regulations, sex-biased cis-eQTM regulations, sex-biased cis-sQTM regulation of each cancer type.
ACC |
BLCA |
BRCA |
CHOL |
COAD |
DLBC |
ESCA |
GBM |
HNSC |
KICH |
KIRC |
KIRP |
LAML |
LGG |
LIHC |
LUAD |
LUSC |
MESO |
PAAD |
PCPG |
READ |
SARC |
SKCM |
STAD |
THCA |
THYM |
UVM |
 Browse the Sex-biased molecules in cancer
Browse the Sex-biased molecules in cancer∗Users can get a list of genes with the sex-biased molecules.
 |
 |
 |
 |
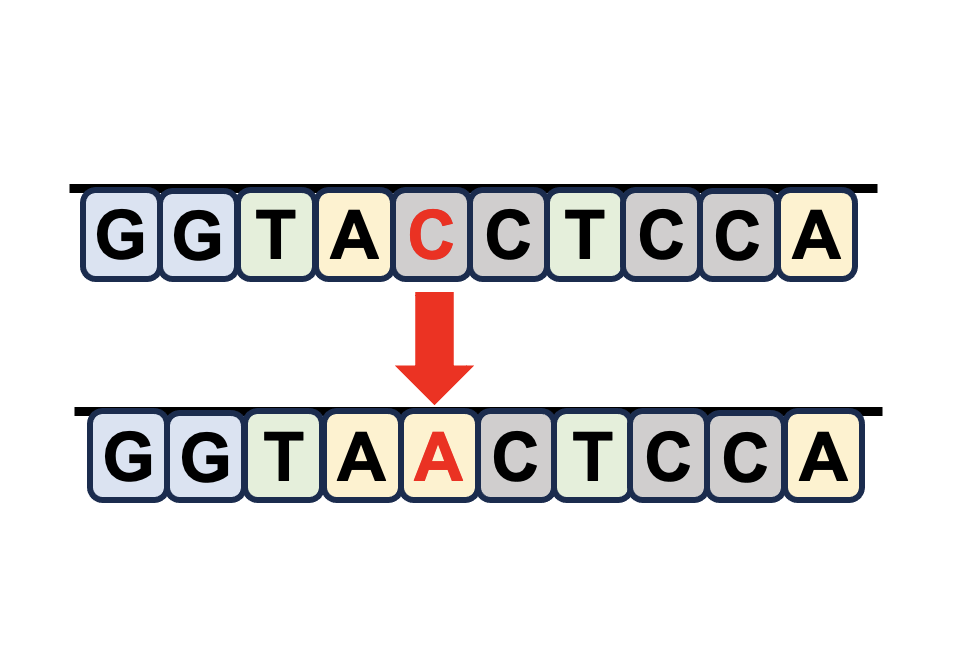
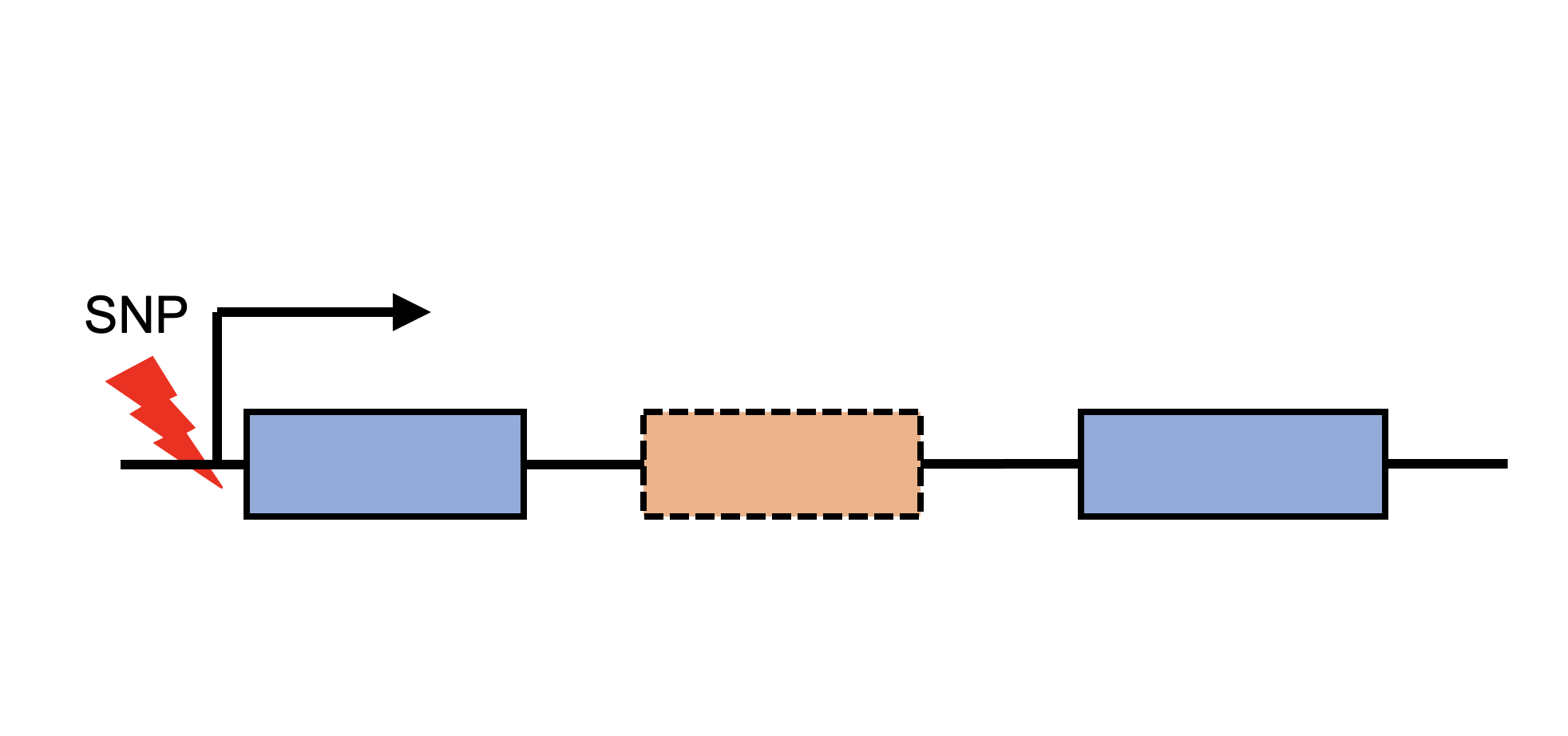
Sex-biased DNA mutations |
Sex-biased mRNAs |
Sex-biased lncRNAs |
Sex-biased exon skipping |
 |
 |
 |
 |
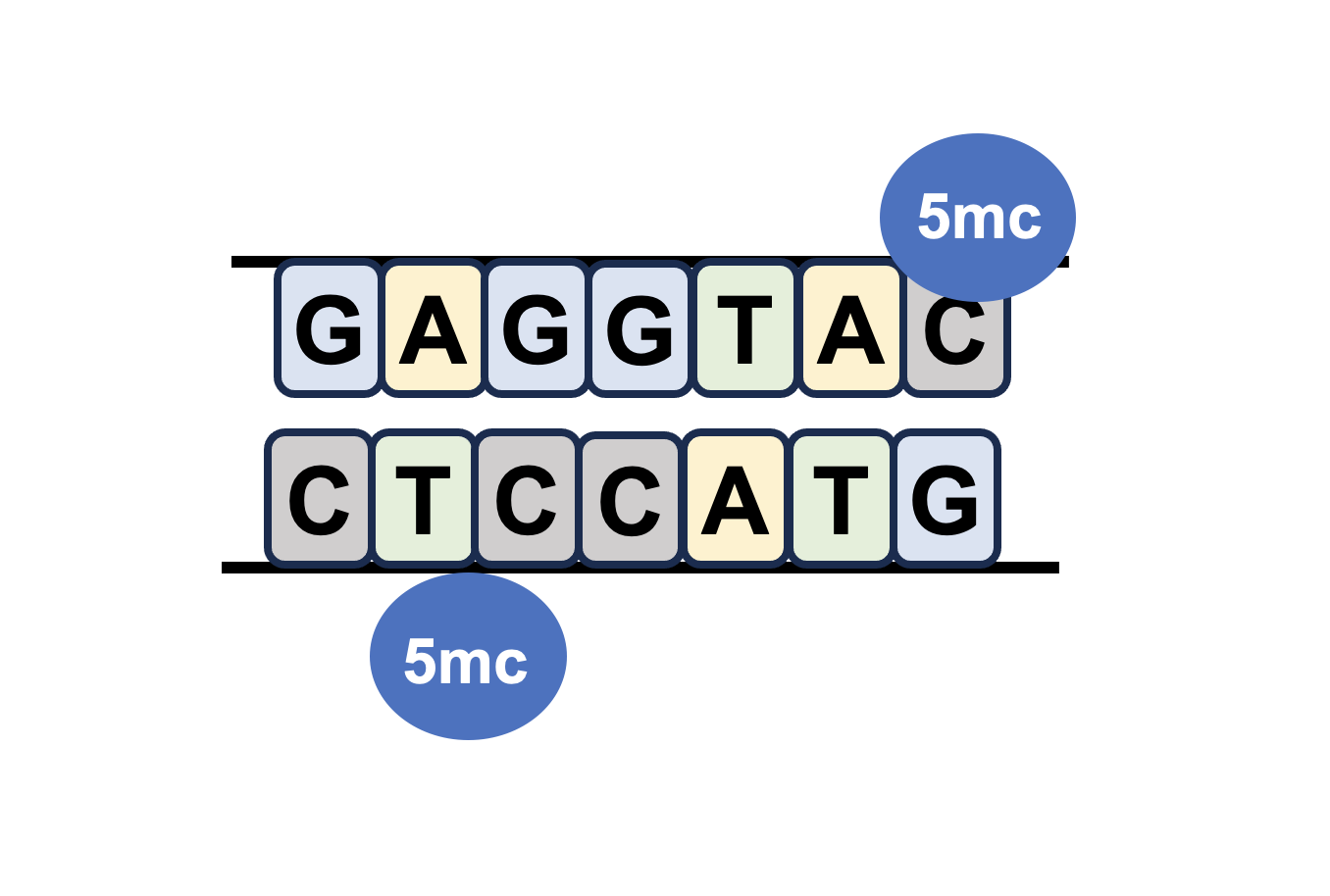
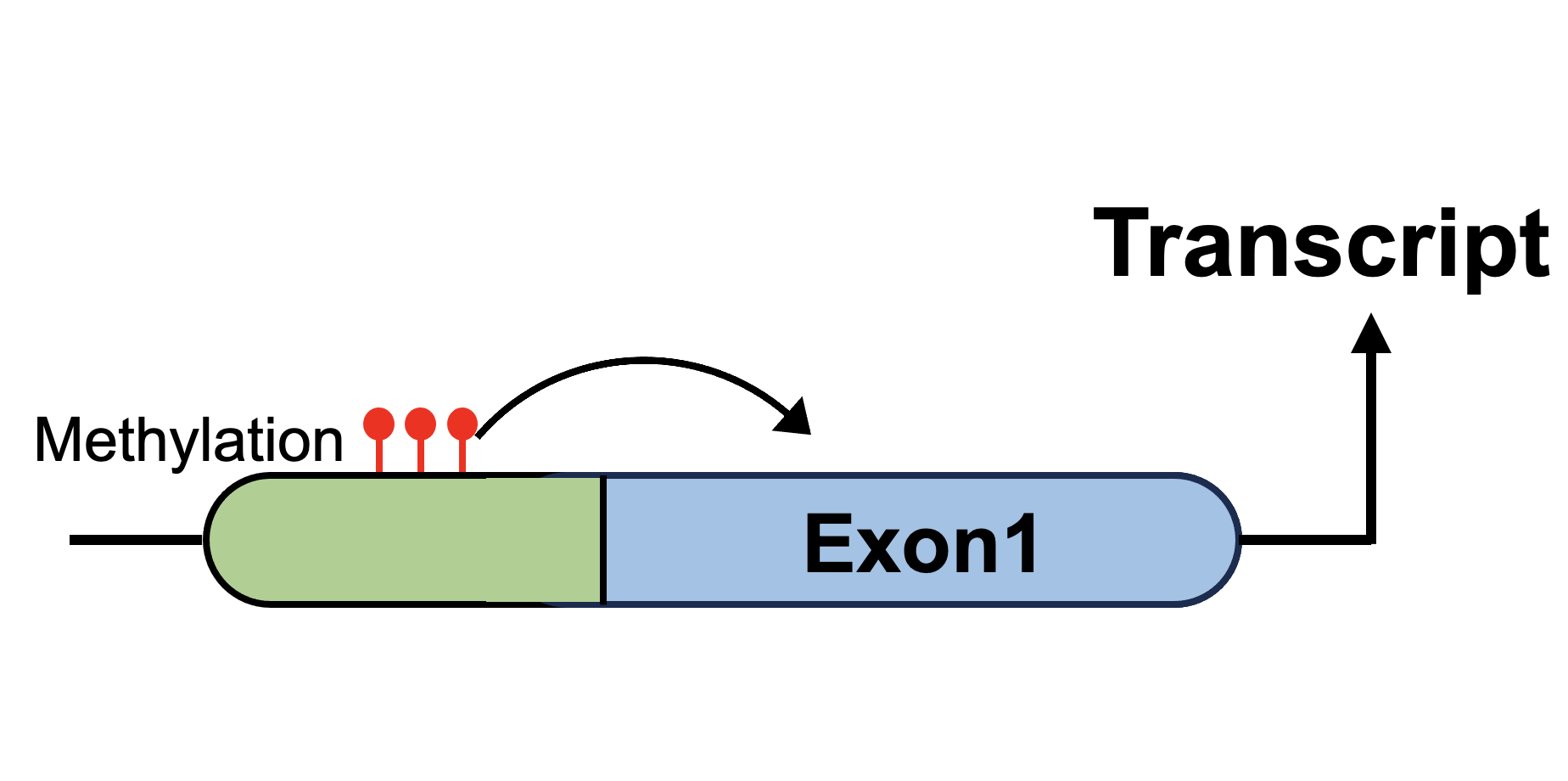
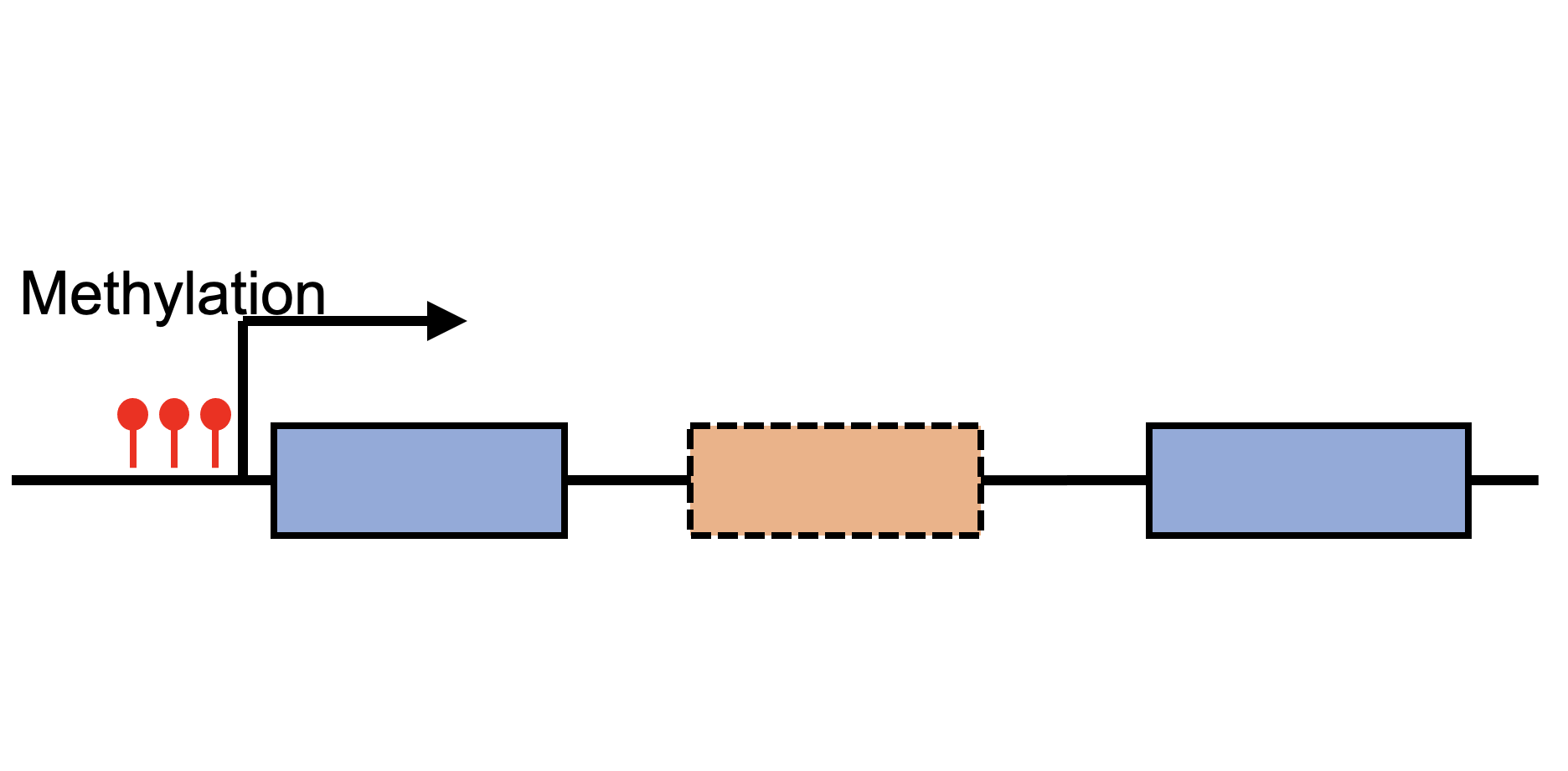
Sex-biased DNA methylations |
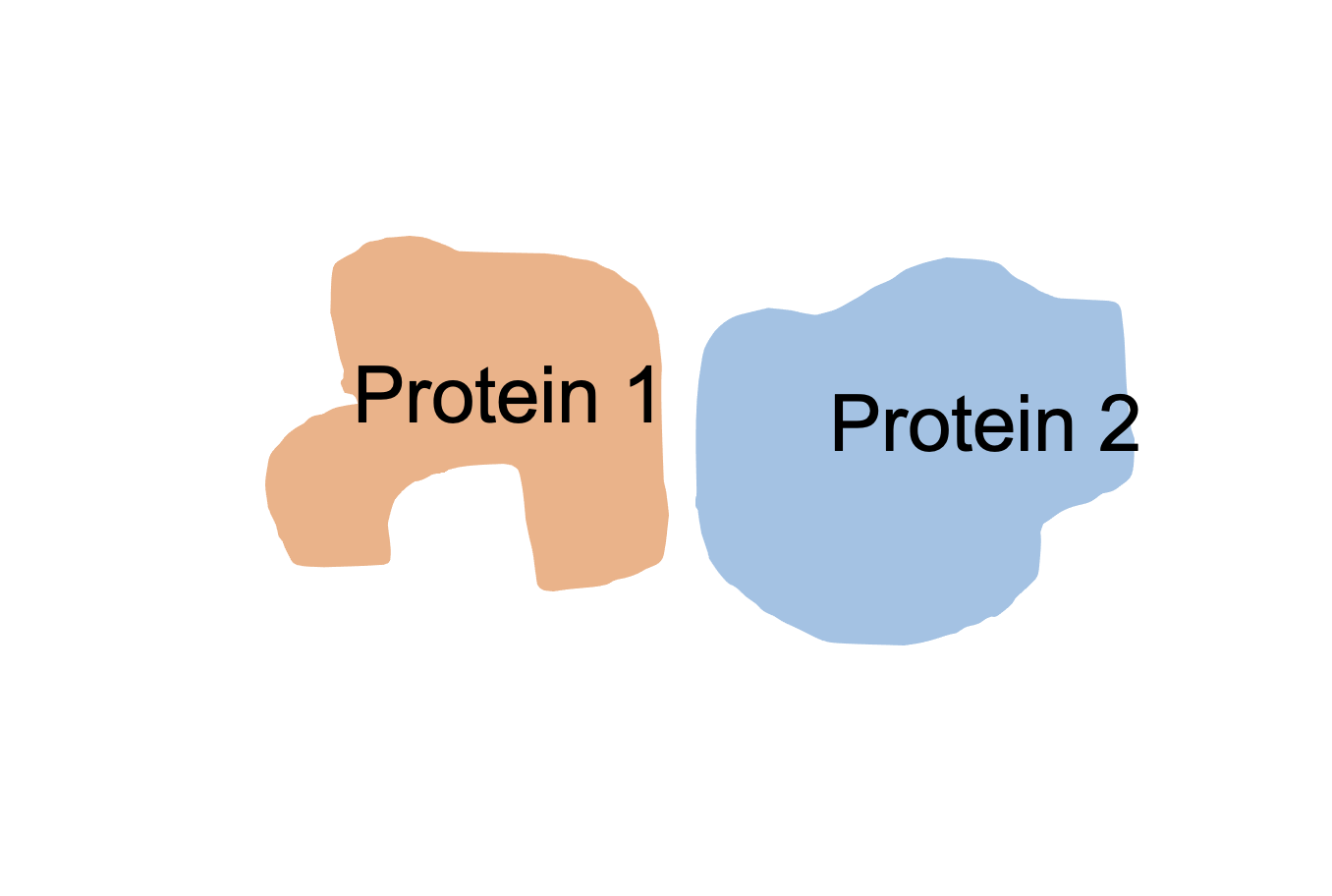
Sex-biased proteins |
Sex-biased miRNAs |
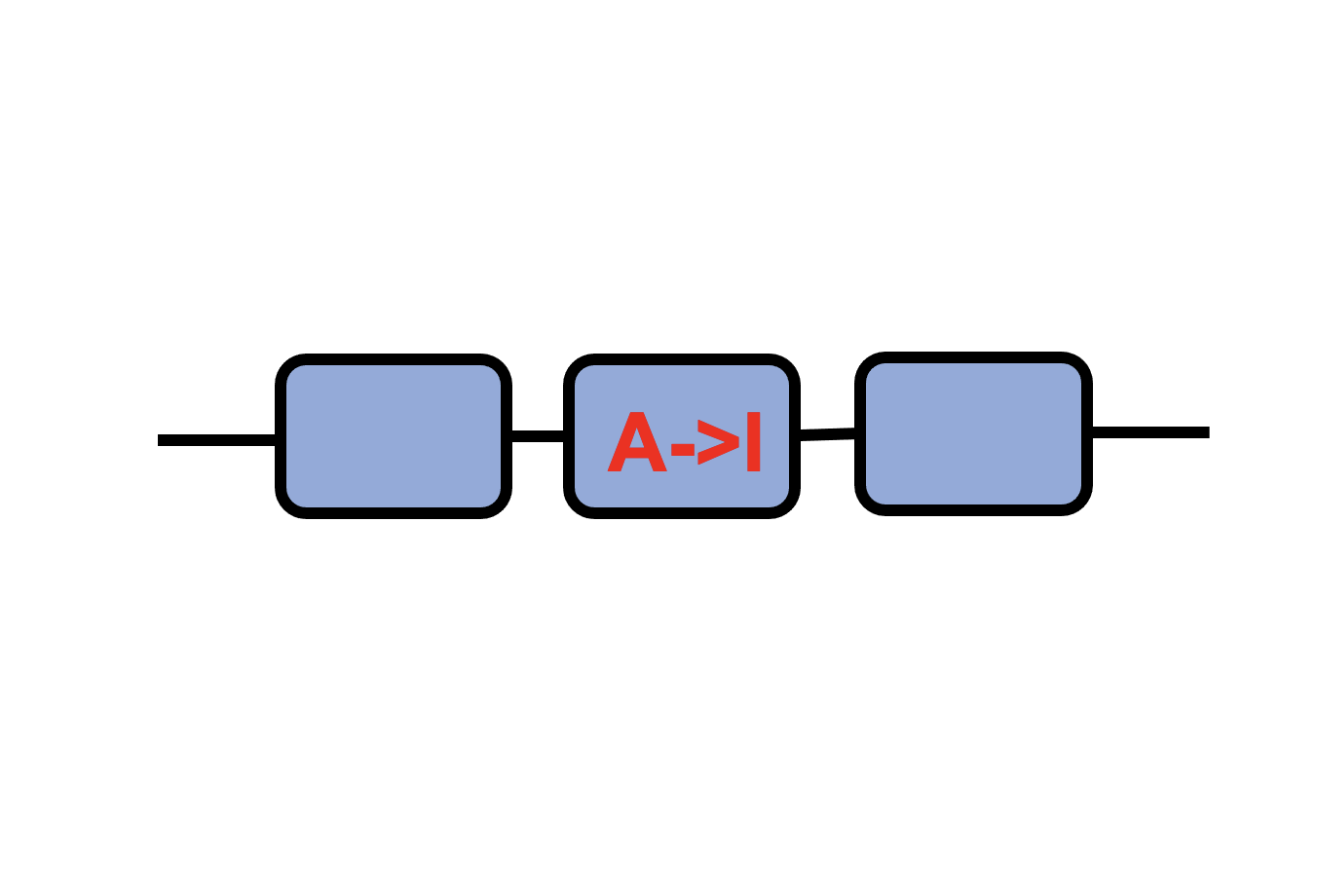
Sex-biased RNA A-to-I editing |
 Sex-biased regulations
Sex-biased regulations∗Users can get a list of sex-biased genes with the corresponding regulations. Then it will be linked to the general information and related regulations of sex-biased genes.
 |
 |
 |
 |
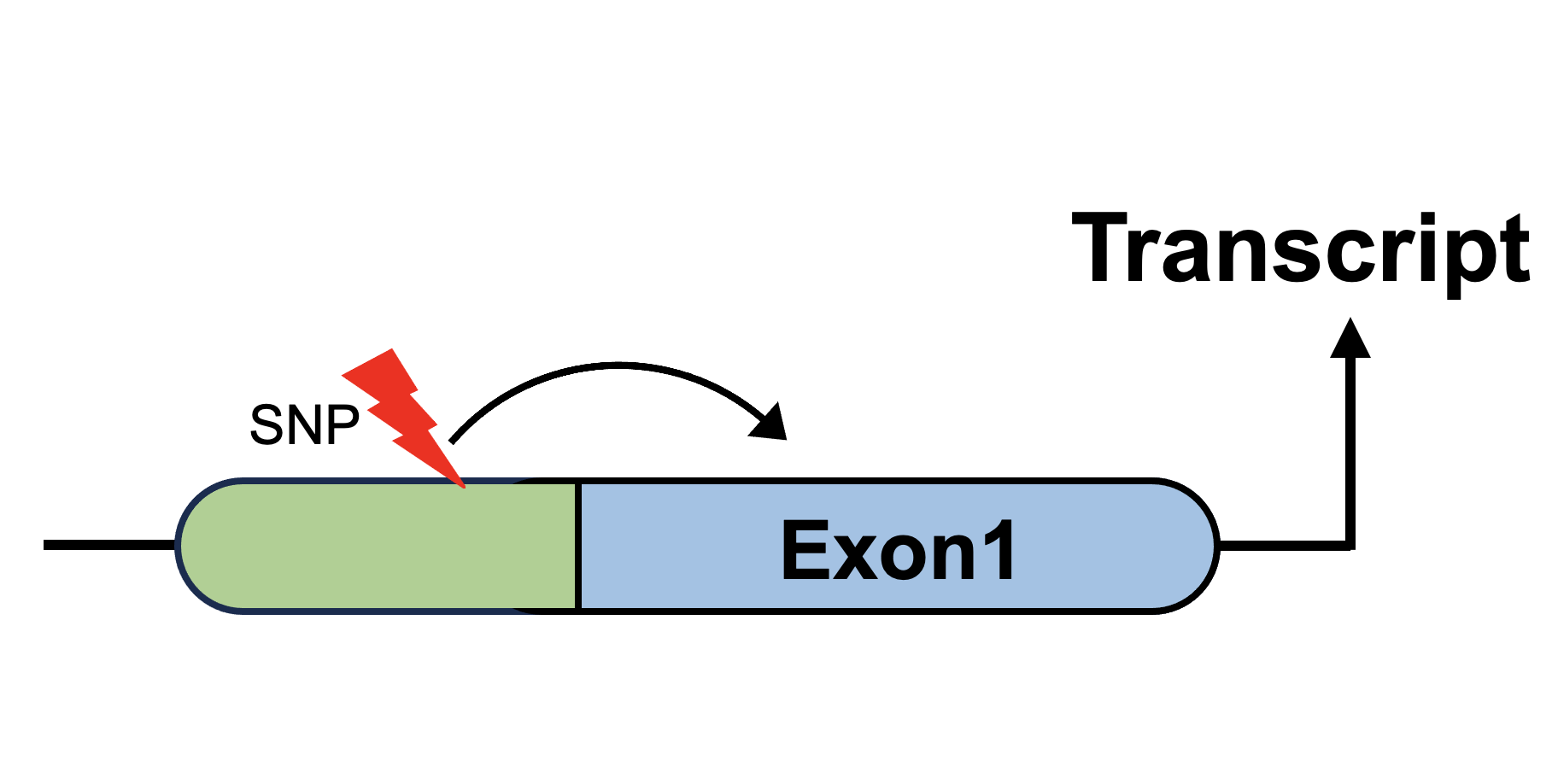
Sex-biased expression Quantitative Trait Loci (eQTL) |
Sex-biased expression Quantitative Trait Methylation (eQTM) |
Sex-biased splicing Quantitative Trait Loci (sQTL) |
Sex-biased splicing Quantitative Trait Methylation (sQTM) |
 |
 |
 |
|
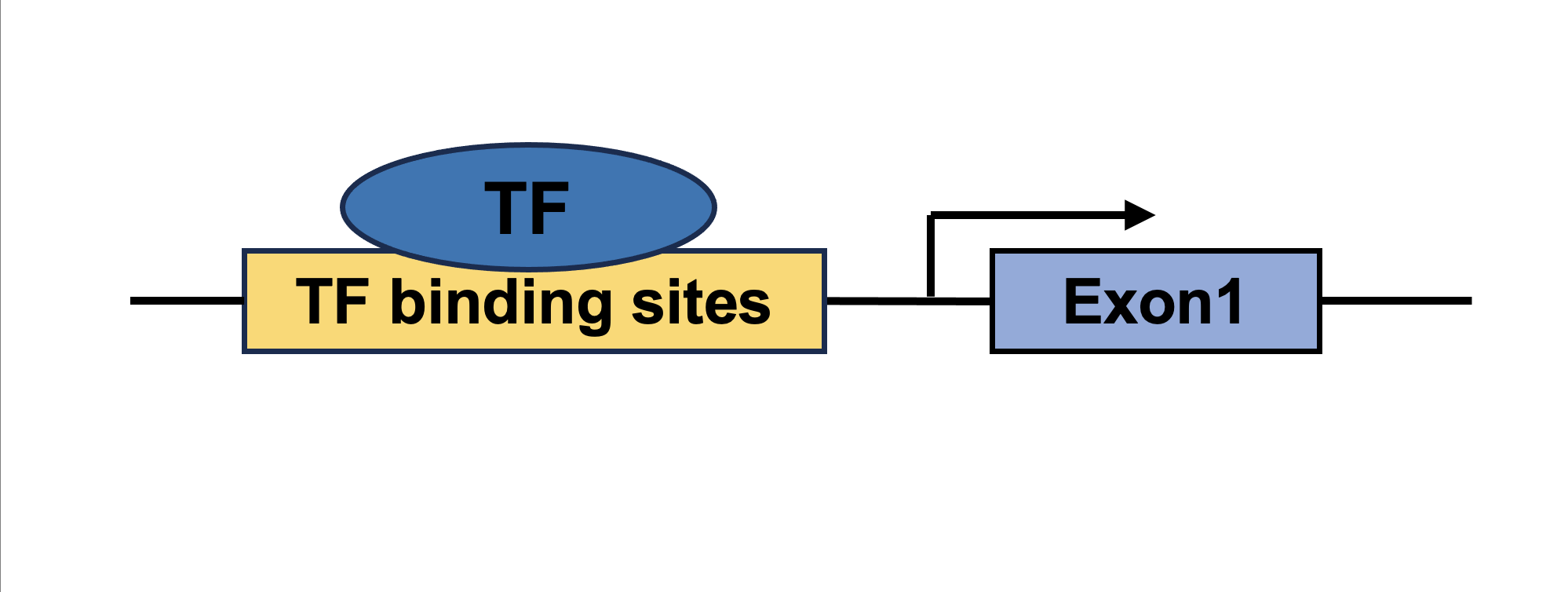
| Sex-biased TF-coding gene network TF | Coding gene |
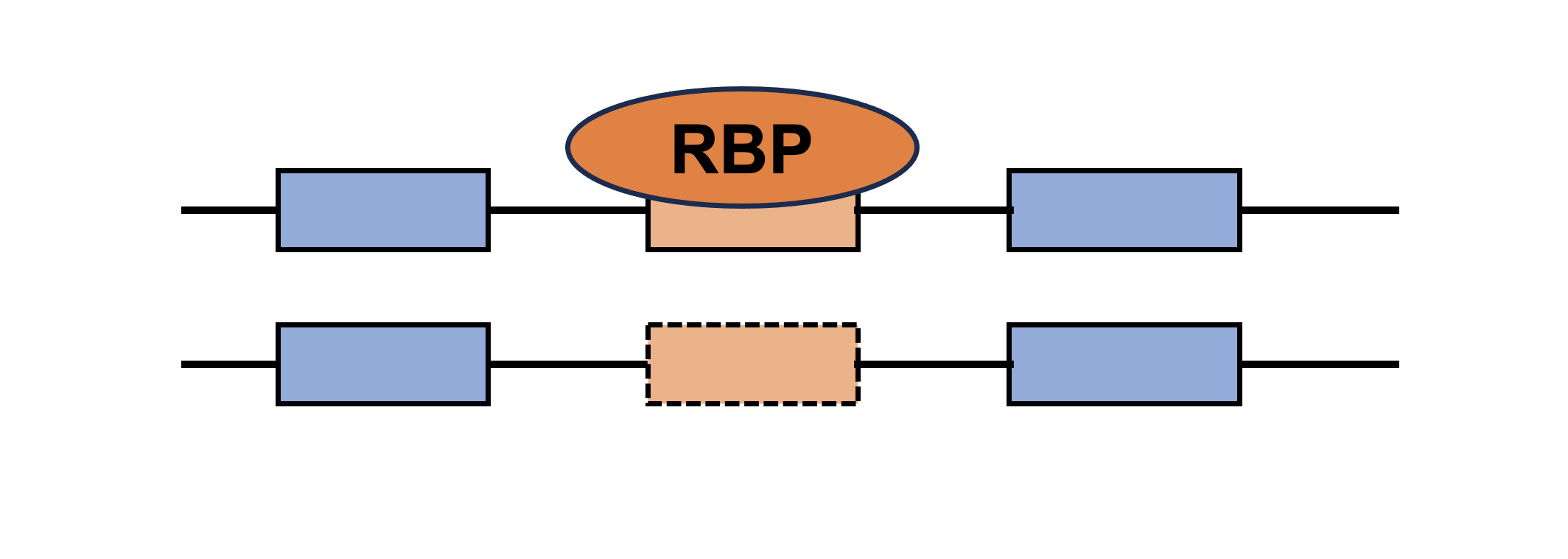
Sex-biased RBP-ES network RBP | Coding gene |
Sex-biased eRNA network |
