
|
||||||
|
||||||
Navigation1.Description of SPASCER2. Understanding annotation of SPASCER from single gene search view Search page, example: DLL1 Gene search result page Gene annotation result page 1) Gene summary 2) Spatial pattern single genes 3) Spatial pattern pathway 4) Cell cell interactions using single cell data 5) Cell cell/spot-spot interactions using spatial data 6) Transcription factors analysis 7) Related drugs and diseases with this gene 3. Understanding annotation of SPASCER from paper search view 1) Search from each paper view 2) Umap of cell types 3) Dotplot of cell type marker genes 4) TFs envolved 5) Heatmap of TFs 6) scRNA-seq cell-cell interactions 7) Cell types distribution using deconvoluted spatial data 8) Cell-cell interactions using deconvoluted spatial data 4. Download data and contact us 1. Spatial database at single cell resolution We We collected 41 datasets, including 1250 samples, are collected, and four species (huamn, mouse, chicken and zebrafish). We conducted spatial single gene pattern, spatial pathway pattern, transcription factor and cell-cell interaction analysis. 2. Understanding SPASCER's annotation categories Search page, example: DLL1 - Input query: Official HUGO gene symbol or Entrez gene ID. 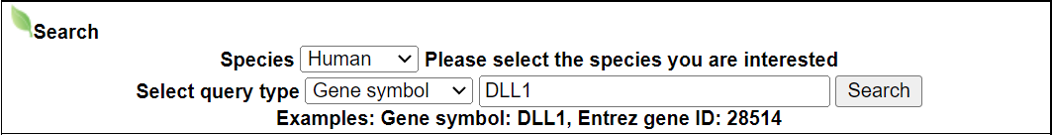 Gene search result page Select your gene from the gene list. 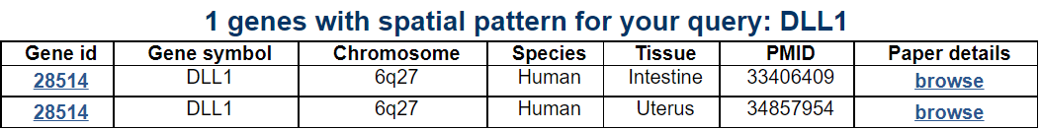 Gene annotation result page These are SPASCER's annotation categories for your query with links to their corresponding annotation parts.  1) Gene summary category This category shows the information of the spatial pattern gene. Firstly, it shows each partner gene's overall information from basic information such as symbol, alias, and locations and ENST accessions. 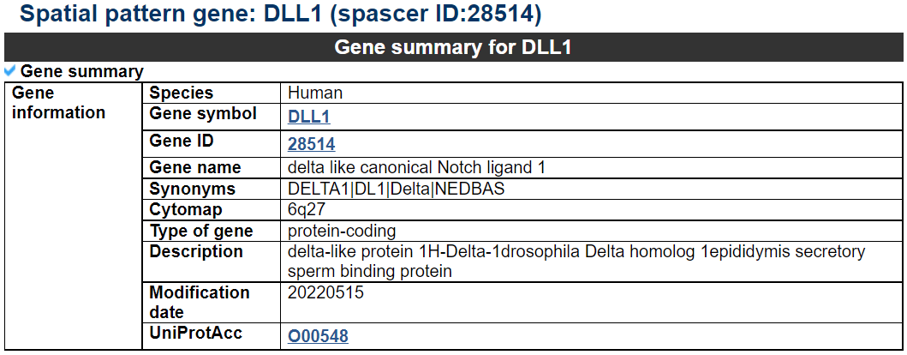
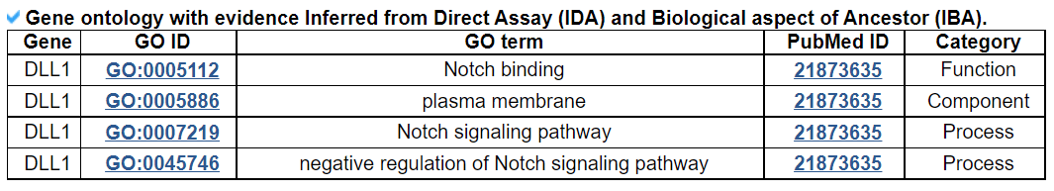
2) 'Single gene with spatial pattern' category This category provides the information whether this gene has spatial heterogeneity across the tissue sample. In the tumor and disease study, if this gene only distributed in the tumor/disease region, it could be a spatial marker for further study. 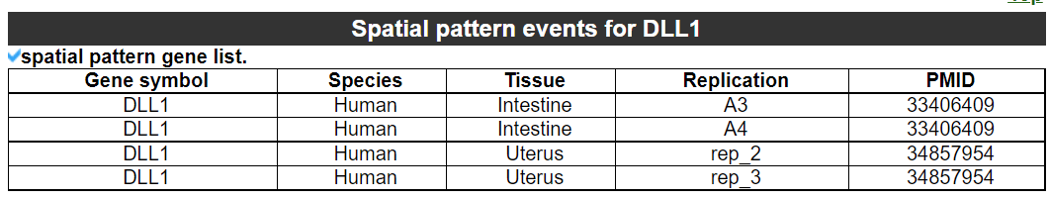 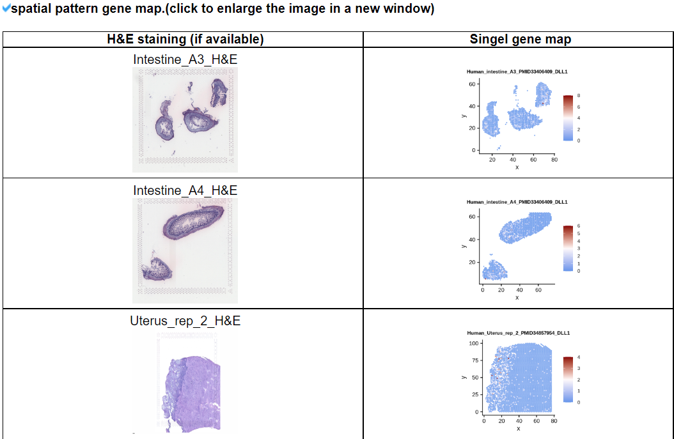 3) 'Spatial pattern pathway' category This category provides the annotation by integrating several genes in a commoon pathway to check whether the pathway is specially expresed in a tissue region. 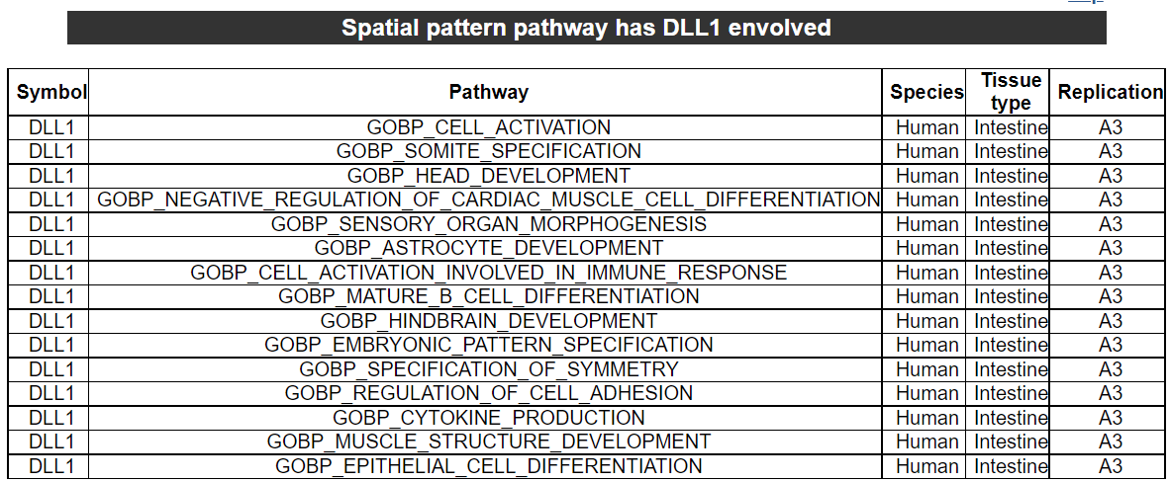 4) 'Cell cell interactions using single cell data' category This category provides the cell cell interaction using single cell data, and provie the statistical significant ligand and receptor pairs.  5) 'Cell cell/spot-spot interactions using spatial data' category This category provides spot-spot interactions interactions and deconvoluted cell-cell interactions across the tissue sample. 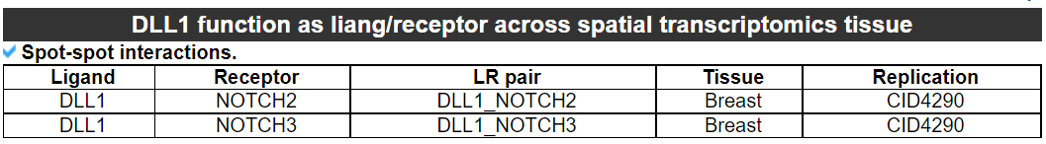 6) 'Transcription factors regulon' category This category provides the transcription factors envolved in two cell types. 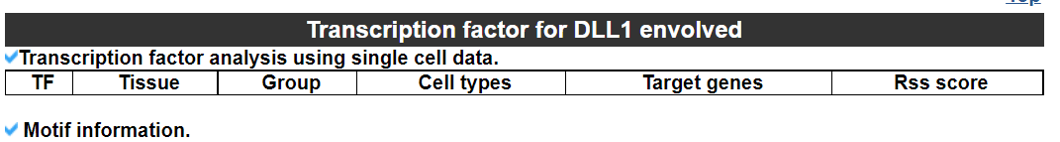 7) 'Related drugs and diseases with this gene' category This table provides the DrugBank information of approved drugs related with spatial pattern gene. Next table provides disease information related to the spatial pattern edited gene.   3. Understanding SPASCER's annotation categories 1) Paper summary This category shows the information about the paper information. 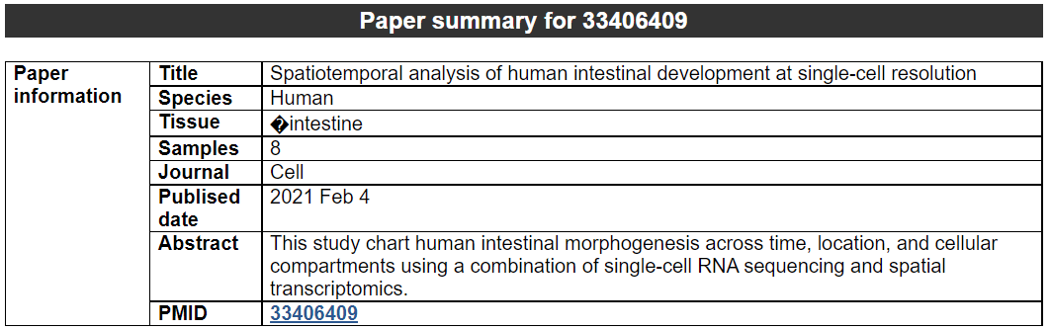 2) Umap of cell types This category shows the information about cell types. 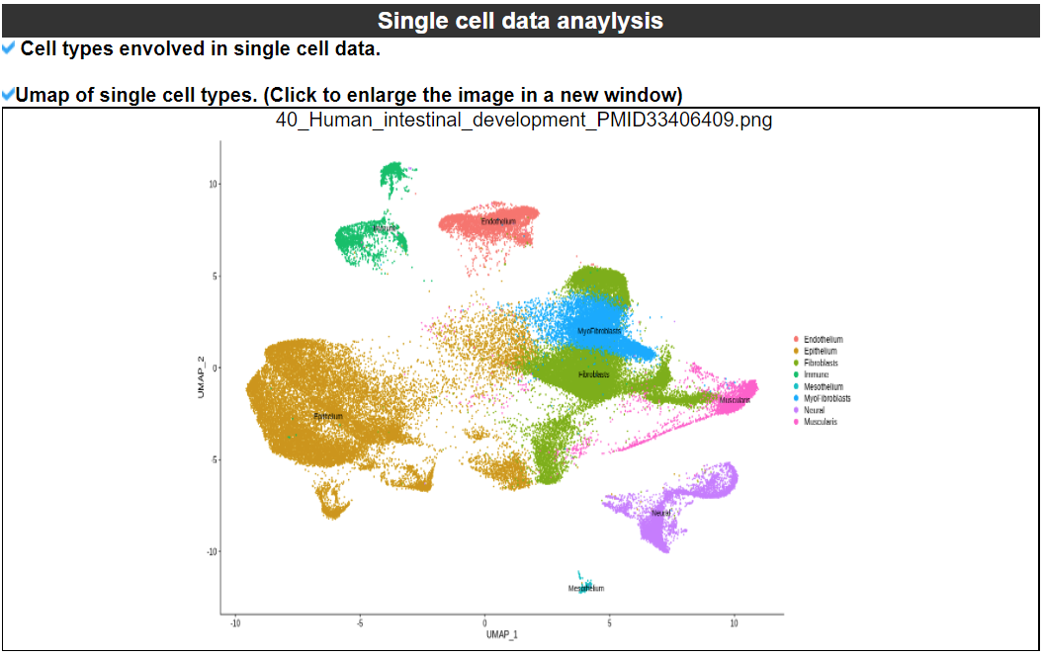 3) scRNA-seq marker genes This category shows the information about cell types marker genes.  4) scRNA-seq TF list This category shows the information about transcription factor envolved in scRNA-seq. 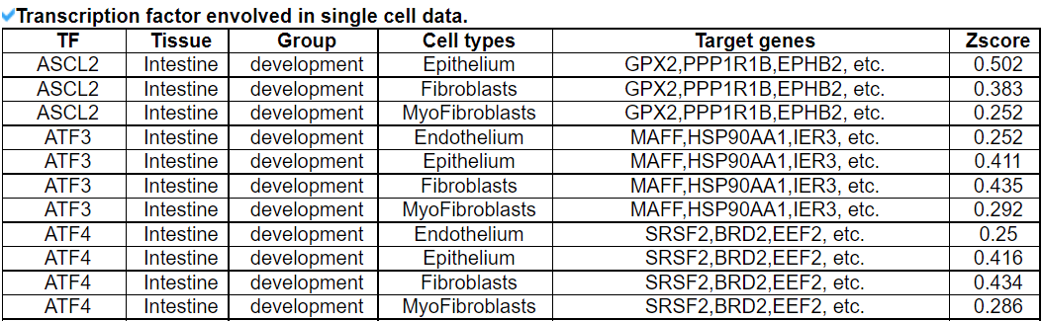 5) Heatmap of TFs This category shows the heatmap about transcription factor envolved in scRNA-seq. 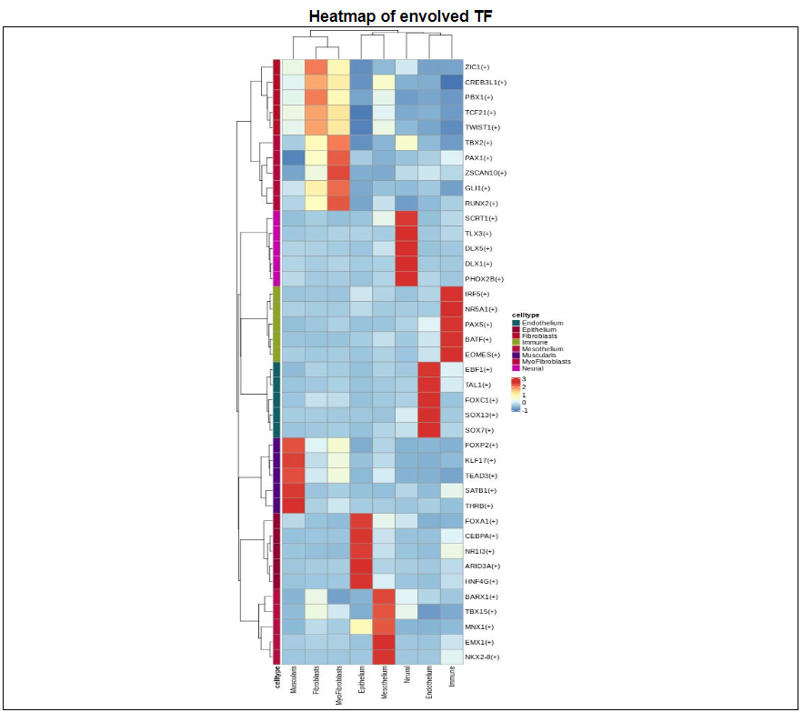 6) scRNA-seq cell-cell interactions This category shows the cell-cell interactions in scRNA-seq. 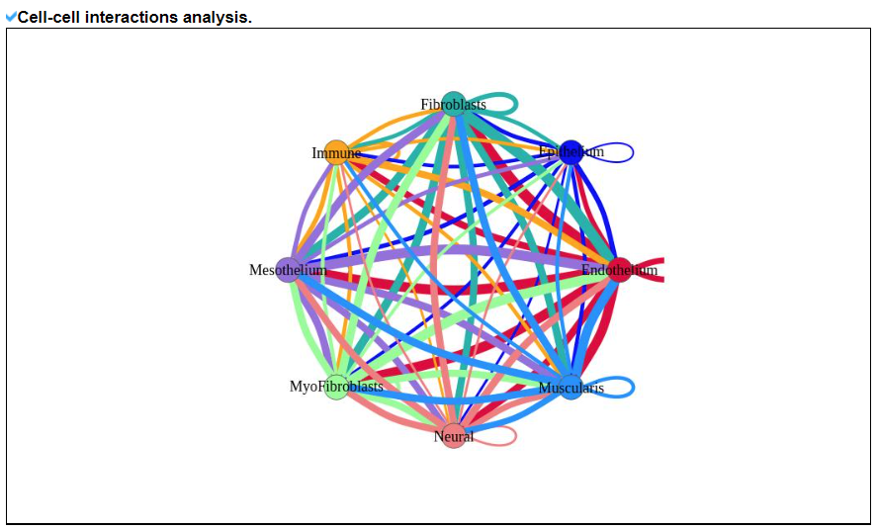 7) Cell types distribution using deconvoluted spatial data This category shows the heatmap about transcription factor envolved in scRNA-seq. 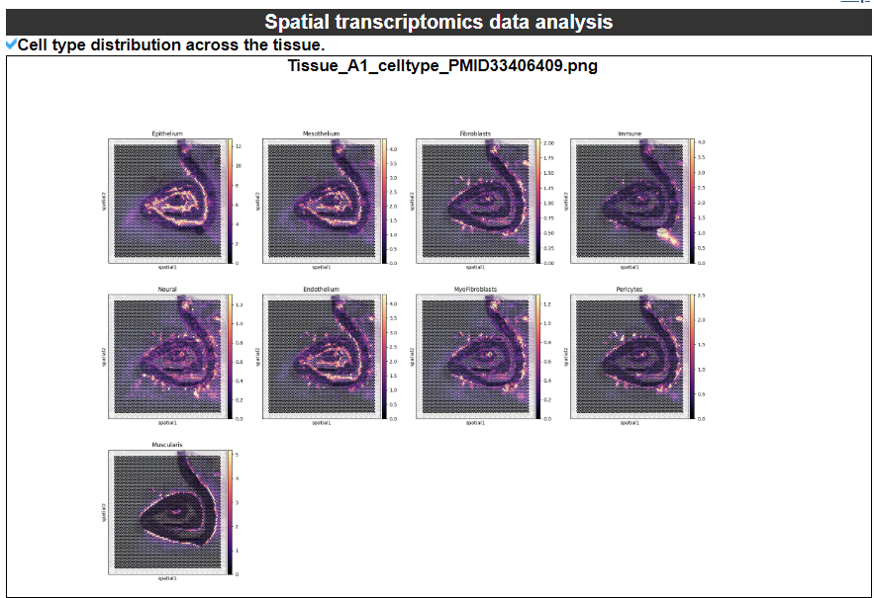 8) cell-cell interactions using deconvoluted spatial data This category shows the heatmap about transcription factor envolved in scRNA-seq. 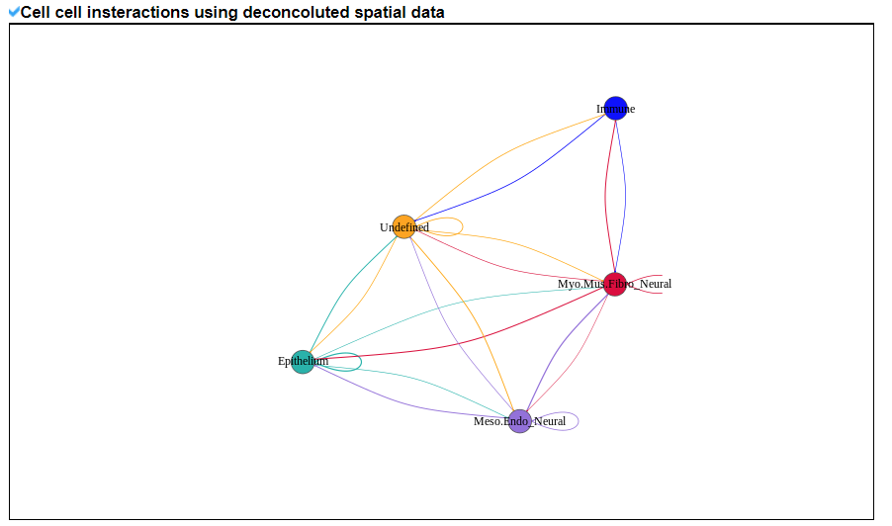 4. Download data and contact us Please go to download page and contact page. |
||||||
