
|
||||||
|
 | |
 | |
 | Differntial gene expression between RNA A-to-I edited tumor samples versus normal samples |
 | |
 | |
 | The effects of the RNA editing to the stability of the RNA structures |
 | |
 | |
 | |
 | |
 |
Editing gene: TRIM25 (CAeditome ID:7706) |
Gene summary for TRIM25 |
 Gene summary Gene summary |
| Gene information | Gene symbol | TRIM25 | Gene ID | 7706 |
| Gene name | tripartite motif containing 25 | |
| Synonyms | EFP|RNF147|Z147|ZNF147 | |
| Cytomap | 17q22 | |
| Type of gene | protein-coding | |
| Description | E3 ubiquitin/ISG15 ligase TRIM25RING finger protein 147RING-type E3 ubiquitin transferase TRIM25estrogen-responsive finger proteintripartite motif protein TRIM25tripartite motif-containing protein 25ubiquitin/ISG15-conjugating enzyme TRIM25zinc fin | |
| Modification date | 20210518 | |
| UniProtAcc | Q14258 | |
| Context |
 Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each this gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Gene | GO ID | GO term | PubMed ID |
| TRIM25 | GO:0003713 | E3 ubiquitin/ISG15 ligase TRIM25 | 23077300 |
| TRIM25 | GO:0043123 | E3 ubiquitin/ISG15 ligase TRIM25 | 23077300 |
| TRIM25 | GO:0043627 | E3 ubiquitin/ISG15 ligase TRIM25 | 22452784 |
| TRIM25 | GO:0045087 | E3 ubiquitin/ISG15 ligase TRIM25 | 18248090 |
| TRIM25 | GO:0046596 | E3 ubiquitin/ISG15 ligase TRIM25 | 18248090 |
| TRIM25 | GO:0005654 | E3 ubiquitin/ISG15 ligase TRIM25 | 0000052 |
| TRIM25 | GO:0005829 | E3 ubiquitin/ISG15 ligase TRIM25 | 0000052 |
| TRIM25 | GO:0010494 | E3 ubiquitin/ISG15 ligase TRIM25 | 23950712 |
| TRIM25 | GO:0016604 | E3 ubiquitin/ISG15 ligase TRIM25 | 0000052 |
Top |
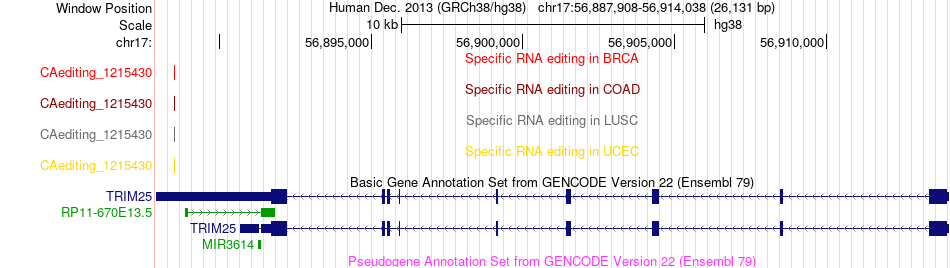
RNA A-to-I events for TRIM25 |
 RNA A-to-I editing events across TCGA 33 cancers data sets based on Ensembl gene isoform structure. RNA A-to-I editing events across TCGA 33 cancers data sets based on Ensembl gene isoform structure. |
 |
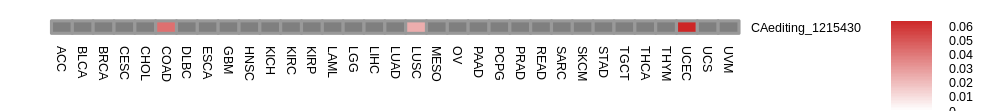
 RNA editing frequencies across 33 cancers of gene: ENSG00000121060.13 RNA editing frequencies across 33 cancers of gene: ENSG00000121060.13 |
 |
 RNA A-to-I editing events in Cancer. RNA A-to-I editing events in Cancer. |
| CAediting_1215430(56888499, -), CAediting_1215431(56894345, -), CAediting_1215432(56894346, -), CAediting_1215433(56894397, -), CAediting_1215434(56896391, -), CAediting_1215435(56896562, -), CAediting_1215436(56896624, -), CAediting_1215437(56896875, -), CAediting_1215438(56896907, -), CAediting_1215439(56896970, -), CAediting_1215440(56897018, -), CAediting_1215441(56905901, -), CAediting_1215442(56905906, -), CAediting_1215443(56905949, -), CAediting_1215444(56906632, -), CAediting_1215445(56906776, -), CAediting_1215446(56911085, -), CAediting_1215447(56911153, -), |
Top |
RNA editing positional annotations for TRIM25 using Annovar |
 Protein coding RNA editing(s). Protein coding RNA editing(s). |
| CAeditome ID | Position | Variant type | ENST | NTchange | AAchange | SIFT_score | Polyphen2_HVAR_score | PROVEAN_pred |
 Gene structure information of RNA editing(s). Gene structure information of RNA editing(s). |
| CAeditome ID | Position | Variant type |
| CAediting_1215430 | chr17_56888499_- | UTR3 |
| CAediting_1215431 | chr17_56894345_- | ncRNA_intronic |
| CAediting_1215432 | chr17_56894346_- | ncRNA_intronic |
| CAediting_1215433 | chr17_56894397_- | ncRNA_intronic |
| CAediting_1215434 | chr17_56896391_- | ncRNA_intronic |
| CAediting_1215435 | chr17_56896562_- | ncRNA_intronic |
| CAediting_1215436 | chr17_56896624_- | ncRNA_intronic |
| CAediting_1215437 | chr17_56896875_- | ncRNA_intronic |
| CAediting_1215438 | chr17_56896907_- | ncRNA_intronic |
| CAediting_1215439 | chr17_56896970_- | ncRNA_intronic |
| CAediting_1215440 | chr17_56897018_- | ncRNA_intronic |
| CAediting_1215441 | chr17_56905901_- | ncRNA_intronic |
| CAediting_1215442 | chr17_56905906_- | ncRNA_intronic |
| CAediting_1215443 | chr17_56905949_- | ncRNA_intronic |
| CAediting_1215444 | chr17_56906632_- | ncRNA_intronic |
| CAediting_1215445 | chr17_56906776_- | ncRNA_intronic |
| CAediting_1215446 | chr17_56911085_- | ncRNA_intronic |
| CAediting_1215447 | chr17_56911153_- | ncRNA_intronic |
 Repat-regional RNA editing(s). Repat-regional RNA editing(s). |
| CAeditome ID | Position | Repeat family | Repeat sub family | Repeat name |
| CAediting_1215431 | chr17_56894345_- | SINE | Alu | AluSz6 |
| CAediting_1215432 | chr17_56894346_- | SINE | Alu | AluSz6 |
| CAediting_1215433 | chr17_56894397_- | SINE | Alu | AluSz6 |
| CAediting_1215434 | chr17_56896391_- | SINE | Alu | AluJb |
| CAediting_1215435 | chr17_56896562_- | SINE | Alu | AluJb |
| CAediting_1215436 | chr17_56896624_- | SINE | Alu | AluJb |
| CAediting_1215437 | chr17_56896875_- | SINE | Alu | AluJr |
| CAediting_1215438 | chr17_56896907_- | SINE | Alu | AluJr |
| CAediting_1215439 | chr17_56896970_- | SINE | Alu | AluJr |
| CAediting_1215440 | chr17_56897018_- | SINE | Alu | AluJr |
| CAediting_1215441 | chr17_56905901_- | SINE | Alu | AluJr |
| CAediting_1215442 | chr17_56905906_- | SINE | Alu | AluJr |
| CAediting_1215443 | chr17_56905949_- | SINE | Alu | AluJr |
| CAediting_1215444 | chr17_56906632_- | SINE | Alu | AluSq2 |
| CAediting_1215445 | chr17_56906776_- | SINE | Alu | AluSq2 |
| CAediting_1215446 | chr17_56911085_- | SINE | Alu | AluJr4 |
| CAediting_1215447 | chr17_56911153_- | SINE | Alu | AluJr4 |
Top |
RNA A-to-I editing events in the alternative splicing sites for TRIM25 |
 RNA A-to-I editing(s) in the alternative splicing sites. RNA A-to-I editing(s) in the alternative splicing sites. |
| AStype | CAeditomID | Editing position | AS position | AS direction | Exonic location | Wildtype sequence | Wildtype splicing strength | RNA edited sequence | RNA edited splicing strength |
 Differential percent of spliced in (PSI) between RNA A-to-I edited and non-edited samples for ENSG00000121060.13.TRIM25. Differential percent of spliced in (PSI) between RNA A-to-I edited and non-edited samples for ENSG00000121060.13.TRIM25. |
 Correlation between RNA A-to-I editing and PSI for ENSG00000121060.13.TRIM25. Correlation between RNA A-to-I editing and PSI for ENSG00000121060.13.TRIM25. |
Top |
Differntial gene expression between RNA A-to-I edited versus non-edited samples for TRIM25 |
 Differential gene expressions between RNA A-to-I edited and non-edited tumor samples. Differential gene expressions between RNA A-to-I edited and non-edited tumor samples.* The grey color means N/A. |
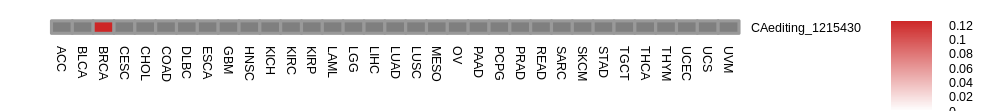 | |
 Correlation between RNA A-to-I editing and gene expression. Correlation between RNA A-to-I editing and gene expression. |
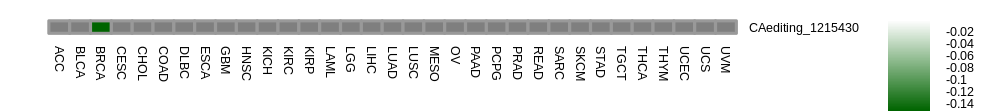 | |
 - Differentially expressed gene between RNA A-to-I edited samples versus non-edited samples. - Differentially expressed gene between RNA A-to-I edited samples versus non-edited samples.* Click on the image to enlarge it in a new window. |
Top |
Protein coding region RNA A-to-I editings for TRIM25 |
 - Lollipop plot for RNA A-to-I editings across protein structure. - Lollipop plot for RNA A-to-I editings across protein structure. * Click on the image to enlarge it in a new window. |
Top |
The effects of the RNA editing to the miRNA binding sites for TRIM25 |
| **If you are searching a miRNA gene with RNA editing events in its seed regions, please see the search pages of miRNA targets to discover the effects of this RNA editing event to miRNA regulations. For more information, please check the files in the Download page. |
 RNA A-to-I editing in the 3'-UTR regions of mRNA gained miRNA binding sites. RNA A-to-I editing in the 3'-UTR regions of mRNA gained miRNA binding sites. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
 RNA A-to-I editing in the 3'-UTR regions of mRNA lost miRNA binding sites. RNA A-to-I editing in the 3'-UTR regions of mRNA lost miRNA binding sites. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
 RNA A-to-I editing in lncRNA gained miRNA binding sites. RNA A-to-I editing in lncRNA gained miRNA binding sites. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
 RNA A-to-I editing in lncRNA lost miRNA binding sites. RNA A-to-I editing in lncRNA lost miRNA binding sites. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
 RNA A-to-I editing in miRNAs gained binding to lncRNA. RNA A-to-I editing in miRNAs gained binding to lncRNA. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
 RNA A-to-I editing in miRNAs lost binding to lncRNA. RNA A-to-I editing in miRNAs lost binding to lncRNA. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
 RNA A-to-I editing in miRNAs gained binding to the 3'-UTR region of mRNA. RNA A-to-I editing in miRNAs gained binding to the 3'-UTR region of mRNA. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
| CAediting_688080 | chr9_63819626_+ | ENSG00000121060.13 | ENST00000572021 | hsa-miR-4477b | 56904334 | 56904340 | 7mer-m8 | 56904333 | 56904356 | 158 | -13.79 |
| CAediting_748296 | chr10_51299626_+ | ENSG00000121060.13 | ENST00000572021 | hsa-miR-605-3p | 56891881 | 56891887 | 7mer-m8 | 56891880 | 56891901 | 140 | -14.43 |
| CAediting_1054808 | chr15_66496985_- | ENSG00000121060.13 | ENST00000316881 | hsa-miR-4512 | 56888810 | 56888817 | 8mer-1a | 56888810 | 56888831 | 140 | -21.39 |
| CAediting_86358 | chr1_109598906_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-197-5p | 56891493 | 56891499 | 7mer-m8 | 56891492 | 56891514 | 150 | -19.91 |
| CAediting_1399425 | chr20_1392958_- | ENSG00000121060.13 | ENST00000316881 | hsa-miR-6869-5p | 56891142 | 56891148 | 7mer-m8 | 56891141 | 56891164 | 152 | -22.81 |
| CAediting_228905 | chr2_188297554_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-561-3p | 56888099 | 56888105 | 7mer-m8 | 56888098 | 56888119 | 140 | -10.96 |
| CAediting_414656 | chr5_72878640_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-4804-3p | 56890662 | 56890668 | 7mer-m8 | 56890661 | 56890684 | 157 | -21.54 |
| CAediting_524911 | chr7_5495852_- | ENSG00000121060.13 | ENST00000316881 | hsa-miR-589-3p | 56889698 | 56889704 | 7mer-m8 | 56889697 | 56889720 | 147 | -19.78 |
| CAediting_573001 | chr7_92204086_- | ENSG00000121060.13 | ENST00000316881 | hsa-miR-1285-5p | 56889496 | 56889502 | 7mer-m8 | 56889495 | 56889513 | 149 | -19.33 |
| CAediting_86358 | chr1_109598906_+ | ENSG00000121060.13 | ENST00000537230 | hsa-miR-197-5p | 56891493 | 56891499 | 7mer-m8 | 56891492 | 56891514 | 150 | -19.91 |
| CAediting_1399425 | chr20_1392958_- | ENSG00000121060.13 | ENST00000537230 | hsa-miR-6869-5p | 56891142 | 56891148 | 7mer-m8 | 56891141 | 56891164 | 152 | -22.81 |
| CAediting_396018 | chr5_1062918_- | ENSG00000121060.13 | ENST00000572021 | hsa-miR-4635 | 56904332 | 56904339 | 8mer-1a | 56904332 | 56904350 | 151 | -15.12 |
| CAediting_1368054 | chr19_46709310_- | ENSG00000121060.13 | ENST00000572021 | hsa-miR-320e | 56895419 | 56895426 | 8mer-1a | 56895419 | 56895436 | 140 | -14.66 |
 RNA A-to-I editing in miRNAs lost binding to the 3'-UTR mRNA. RNA A-to-I editing in miRNAs lost binding to the 3'-UTR mRNA. |
| CAeditom_ID | Position | ENSG | ENST | miRNA | Targetscan start | Targetscan end | Targetscan score | Miranda start | Miranda end | Miranda score | Miranda energy |
| CAediting_77128 | chr1_85133844_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-4423-3p | 56891482 | 56891488 | 7mer-m8 | 56891481 | 56891501 | 150 | -16.49 |
| CAediting_1229414 | chr17_67471546_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-548aa | 56891045 | 56891051 | 7mer-m8 | 56891044 | 56891069 | 151 | -9.3 |
| CAediting_1229415 | chr17_67471547_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-548aa | 56891045 | 56891051 | 7mer-m8 | 56891044 | 56891069 | 151 | -9.3 |
| CAediting_117844 | chr1_162342605_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-556-3p | 56888320 | 56888326 | 7mer-m8 | 56888319 | 56888340 | 142 | -7.31 |
| CAediting_228905 | chr2_188297554_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-561-3p | 56891030 | 56891036 | 7mer-m8 | 56891029 | 56891048 | 153 | -6.38 |
| CAediting_524911 | chr7_5495852_- | ENSG00000121060.13 | ENST00000316881 | hsa-miR-589-3p | 56891449 | 56891455 | 7mer-m8 | 56891448 | 56891471 | 149 | -13.82 |
| CAediting_748296 | chr10_51299626_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-605-3p | 56890756 | 56890762 | 7mer-m8 | 56890755 | 56890776 | 141 | -14.71 |
| CAediting_980496 | chr14_31014685_- | ENSG00000121060.13 | ENST00000316881 | hsa-miR-624-3p | 56889777 | 56889782 | 6mer | 56889776 | 56889796 | 146 | -10.64 |
| CAediting_1351755 | chr19_40282625_- | ENSG00000121060.13 | ENST00000316881 | hsa-miR-641 | 56887964 | 56887969 | 6mer | 56887963 | 56887986 | 151 | -20.79 |
| CAediting_580943 | chr7_100356700_+ | ENSG00000121060.13 | ENST00000316881 | hsa-miR-6840-3p | 56888356 | 56888363 | 8mer-1a | 56888356 | 56888378 | 143 | -24.91 |
| CAediting_524911 | chr7_5495852_- | ENSG00000121060.13 | ENST00000537230 | hsa-miR-589-3p | 56891449 | 56891455 | 7mer-m8 | 56891448 | 56891471 | 149 | -13.82 |
| CAediting_228905 | chr2_188297554_+ | ENSG00000121060.13 | ENST00000537230 | hsa-miR-561-3p | 56891030 | 56891036 | 7mer-m8 | 56891029 | 56891048 | 153 | -6.38 |
 Differentially expressed gene-miRNA network Differentially expressed gene-miRNA network |
Top |
The effects of the RNA editing to the stability of the RNA structures for TRIM25 |
 - RNA A-to-I editing in mRNA. - RNA A-to-I editing in mRNA.* Click on the image to enlarge it in a new window. |
| ENST | ENST type | RNA structure without RNA A-to-I editing. | RNA structure with RNA A-to-I editing. |
 RNA A-to-I editing in RNA structures for Minimum free energy (MFE). RNA A-to-I editing in RNA structures for Minimum free energy (MFE). |
| CAeditom_ID | ENST | Editing_Position | MFE_withoneAI | MFE_withoutAI | MFE_withmultipleAI |
Top |
Relation with ADAR for TRIM25 |
 Correlation between ADAR gene expression and RNA editing frequency Correlation between ADAR gene expression and RNA editing frequency |
| Cancer | CAeditomID | position | ADAR1 (p-val) | ADAR1 (coeff.) | ADAR2 (p-val) | ADAR2 (coeff.) | ADAR3 (p-val) | ADAR3 (coeff.) |
| LUAD | CAediting_1215430 | chr17_56888499_- | . | . | 0.0353424108447206 | 0.243417240206009 | . | . |
Top |
Relation with cancer stages for TRIM25 |
 Correlation between Cancer stages and RNA editing frequency Correlation between Cancer stages and RNA editing frequency |
| Cancer | Stage_type | CAeditomID | position | P-val | Coeff. |
| BRCA | pathologic_stage | CAediting_1215430 | chr17_56888499_- | 0.0222978496813893 | -0.140350049610173 |
Top |
Relation with survival for TRIM25 |
 Correlation between RNA editing and survival Correlation between RNA editing and survival |
| Cancer | CAeditomID | position | KMpvalue | Coxpvalue | CoxHR |
| UCEC | CAediting_1215430 | chr17_56888499_- | 0.0276096008573378 | 0.0245541 | 425766 |
Top |
RelatedDrugs for TRIM25 |
 Approved drugs targeting this gene. Approved drugs targeting this gene. (DrugBank Version 5.1.8 2021-01-03) |
| Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
RelatedDiseases for TRIM25 |
 Diseases associated with this gene. Diseases associated with this gene. (DisGeNet 7.0) |
| Gene | Disease ID | Disease name | # pubmeds | Source |
